News
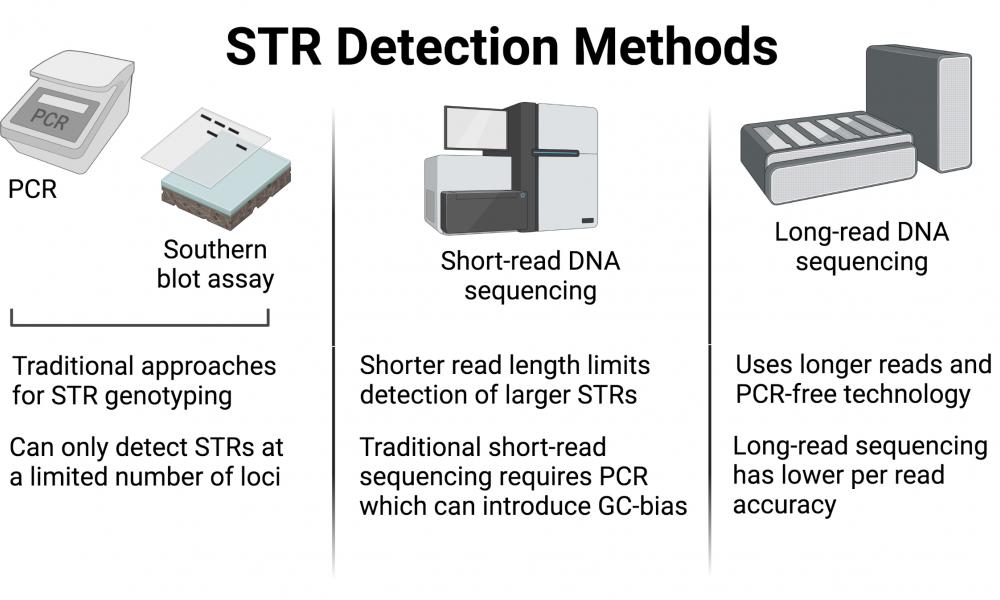
Straglr: a new software tool for targeted genotyping and repeat expansion detection
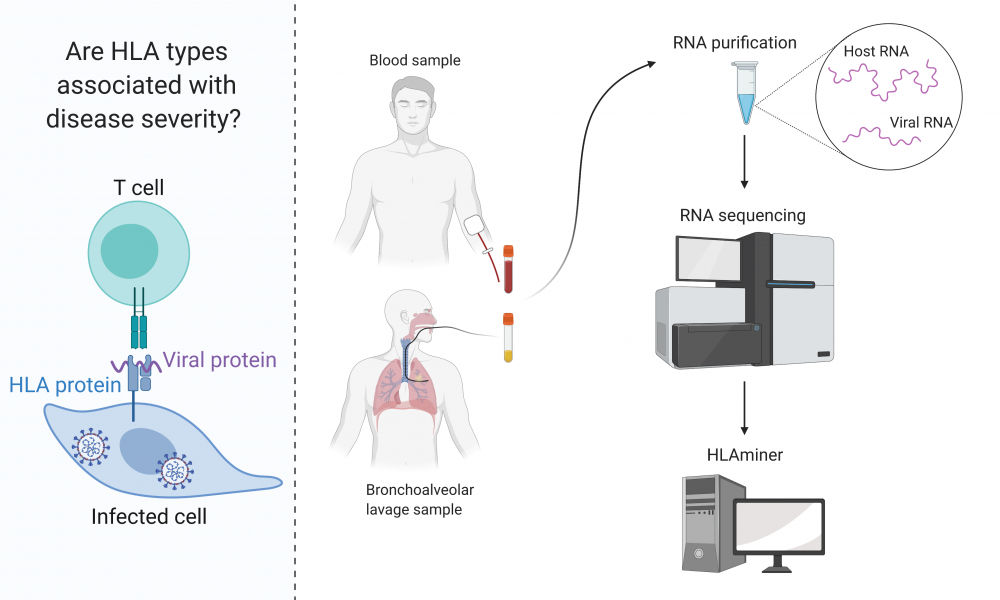
A bioinformatics method may help uncover link between immune system variability and SARS-CoV-2 susceptibility
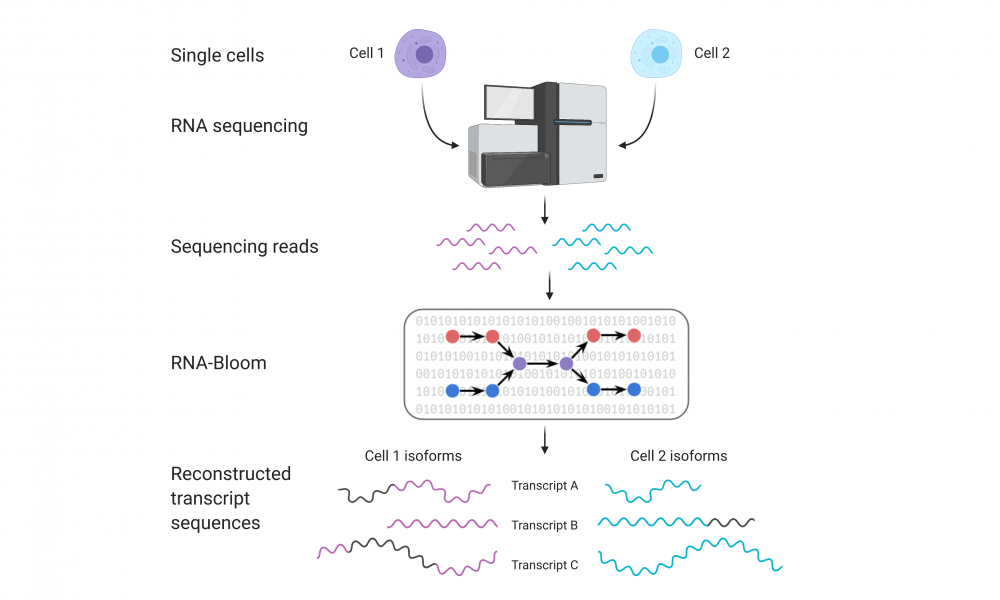
New tool enables researchers to assemble transcript isoforms from single cells
The advent of single-cell RNA sequencing technologies has provided unprecedented opportunities for the analysis of transcriptomes at single-cell resolution, allowing researchers to explore cell-to-cell variability. Now, researchers have developed a tool for the analysis and identification of RNA isoforms from single-cell RNA sequencing data.
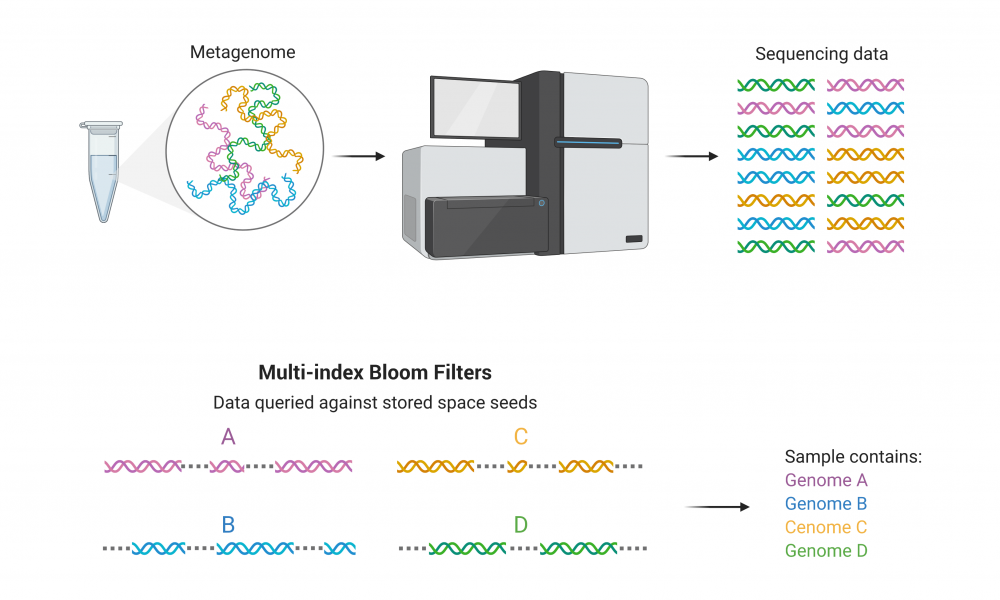
A novel tool for simultaneous classification of DNA from multiple species
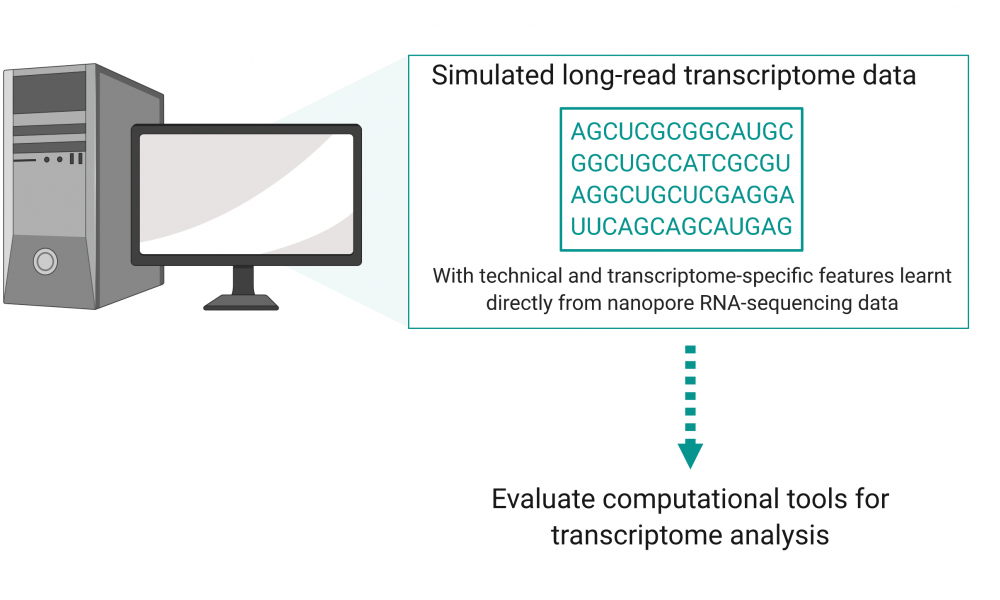
New sequence simulator helps leverage power of long-read transcriptome sequencing
Long-read sequencing technologies are increasingly being employed by researchers to gain important insights into the transcriptomes of cells, revealing a need for computational tools designed for long-read RNA sequencing analysis. To facilitate software development, researchers have now created a sequence simulator designed to produce simulated long-read transcriptome data, providing a cost-effective means to help develop, refine and benchmark novel tools for data analysis.
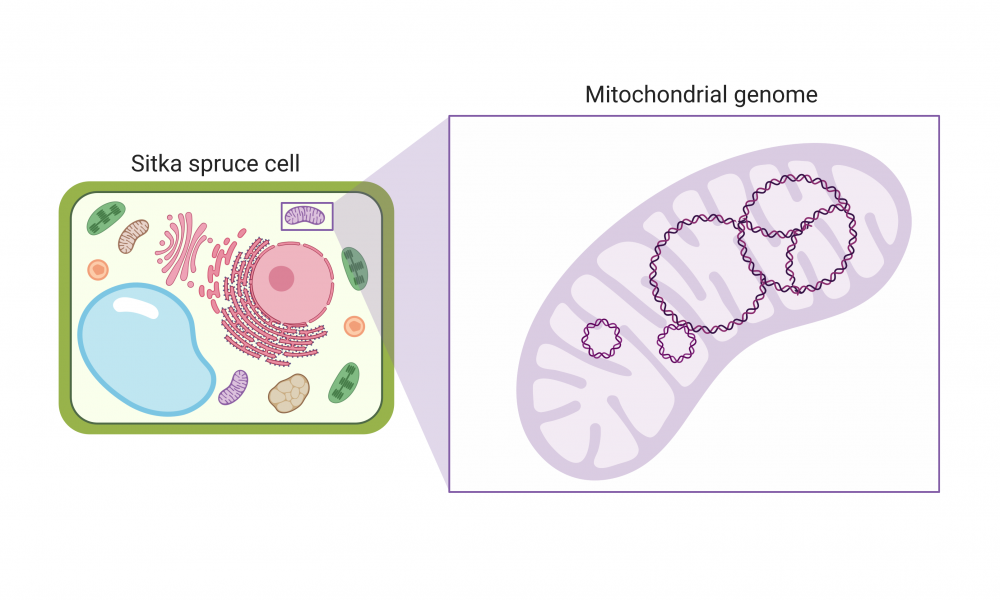
GSC scientists uncover the complex physical structure of the Sitka spruce mitochondrial genome
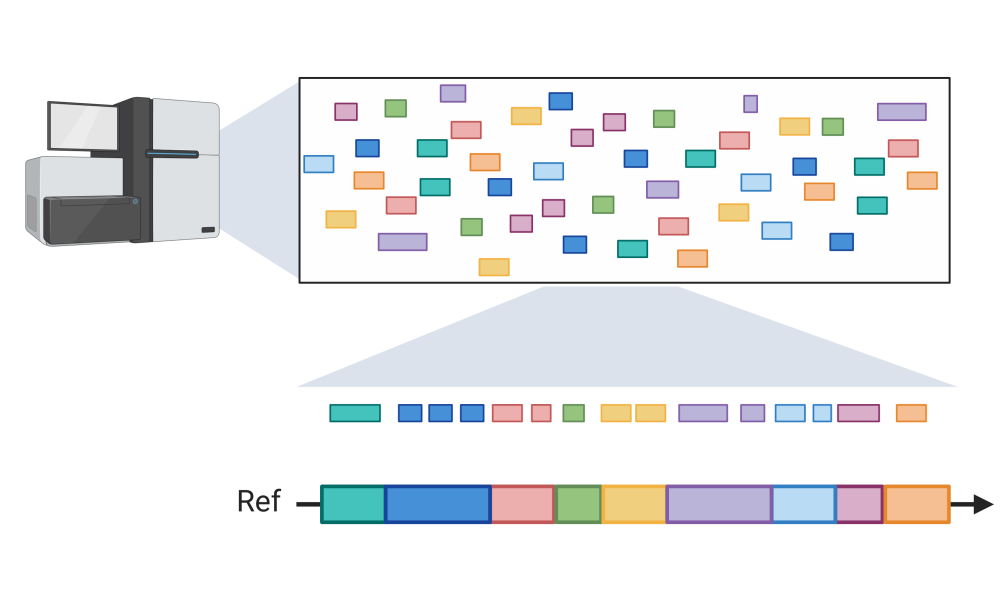
The long and the short of it: scientists develop a tool for rapid genome assembly

Genome British Columbia, LSARP grant: Developing alternatives to using antibiotics in farming
Dr. Inanc Birol was awarded $6.9 million for a project that will employ genomics research to discover and develop antimicrobial peptides (AMPs) as alternatives to traditional antibiotics. Treating and preventing bacterial infections in animals is an essential part of agriculture. However, overuse and misuse of antibiotics has led to the development of antimicrobial resistance, an issue classified by the World Health Organization as one of the most urgent global health risks facing us today.
