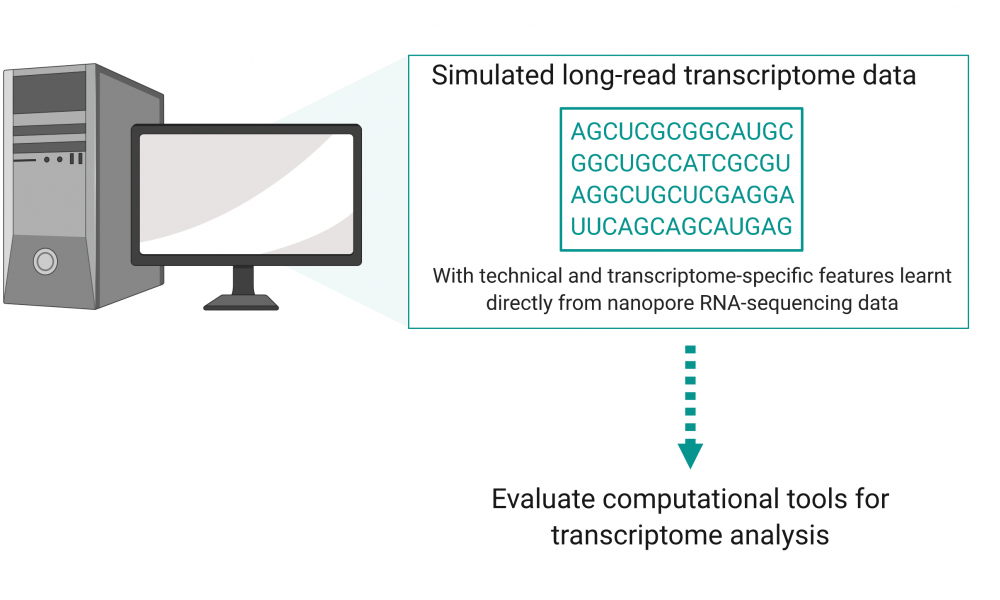
Long-read sequencing technologies are increasingly being employed by researchers to gain important insights into the transcriptomes of cells, revealing a need for computational tools designed for long-read RNA sequencing analysis. To facilitate software development, researchers have now created a sequence simulator designed to produce simulated long-read transcriptome data, providing a cost-effective means to help develop, refine and benchmark novel tools for data analysis.

Transcriptome analysis using long-read sequencing technologies, such as those developed by Oxford Nanopore Technologies, has several advantages over the use of short-read next generation RNA sequencing. Critically, it allows researchers to sequence the full length of transcripts rather than producing RNA sequences in short fragments. This enables detection of alternative splicing, RNA modification and complex features.
Now, in a study published in GigaScience led by GSC Distinguished Scientist Dr. Inanc Birol, researchers describe the development of a new sequence simulator—Trans-NanoSim—specifically designed to simulate transcriptomic data. Simulated data with a known ground truth will enable scientists to develop the computational tools needed to leverage the power of long-read transcriptome data in a cost-effective way.
Dr. Birol’s group previously developed NanoSim—a tool that simulate long-read genomic data. The group took a similar approach to create Trans-NanoSim to simulate reads with technical and transcriptome-specific features learnt directly from real nanopore RNA-sequencing data.
NanoSim and Trans-NanoSim, along with an extensive collection of software packages developed in-house at the GSC, are freely available here and through GitHub.
Learn more about Software Development.
Learn more about Sequencing Services.
Learn more about Bioinformatics Services.