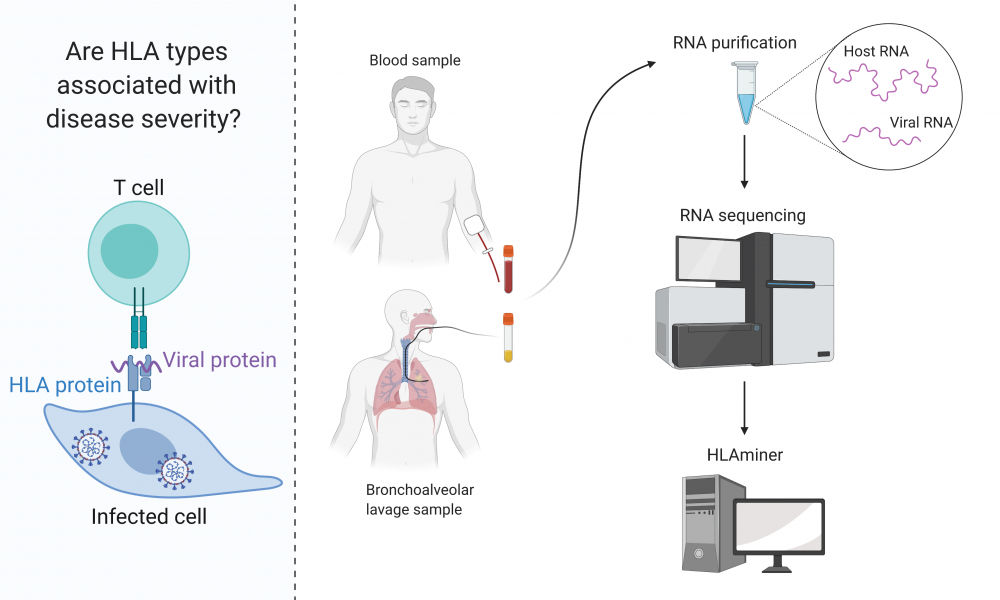
The striking variability in disease severity in people infected with SARS-CoV-2 has been attributed to several host factors, including age, pre-existing health conditions and differences in host genetics and immune responses. As researchers around the world work to shed light on host factors impacting COVID-19 disease, novel tools that enable scientists to mine large datasets are needed.
In a study published in the journal Bioinformatics, GSC Distinguished Scientist Dr. Inanc Birol and group leader René Warren demonstrated the technical feasibility and utility of their bioinformatics tool, HLAminer, to investigate the role of host immune system variability in COVID-19 disease susceptibility.
Human leukocyte antigen (HLA) proteins are essential components of the human immune response to infectious disease. HLA proteins are present on the surface of cells, and they are responsible for telling the immune system whether or not an infection is present.
There are two classes of HLA proteins. If a cell engulfs a virus or becomes infected by one, HLA class II or HLA class I proteins, respectively, will display viral peptides on the cell surface. Specialized immune cells, called T cells, will then detect these viral peptides and activate an immune response.
In the human population, there is a large degree of variability in both classes of HLA, and different HLA alleles have been shown to impact disease outcomes. To enable researchers to explore the link between HLA alleles and disease, Warren had developed a bioinformatics tool called HLAminer in 2012. The tool, which is available for download on GitHub, mines high-throughput next-generation sequencing data.
“Because of the central role of HLA in host immunity and reported associations with disease susceptibility, knowledge about the HLA profiles of COVID-19 patients and asymptomatic SARS-CoV-2 positive individuals could help inform public health response strategies,” says Warren.
In their new study, Warren demonstrated that HLAminer can be used to determine HLA types from both bronchoalveolar lavage and blood samples taken from patients diagnosed with COVID-19. RNA was extracted from eight patient samples, and HLAminer was used to analyze RNA-sequencing data, successfully identifying HLA alleles for all samples.
While the analysis of HLA types in this study uncovered some intriguing findings, the number of patients studied was very small and further analyses are needed to determine the impact of HLA type on disease susceptibility and severity. This study provides an effective method that will enable researchers to enhance our understanding of the interplay between host immune factors and infectious diseases, including SARS-CoV-2.
“We hope that our report will sensitize scientists and clinicians to the importance of HLA determination and the practicality of deriving this information from COVID-19 clinical samples using RNA-sequencing, likely readily available and from sizeable cohorts,” says Warren.
René Warren and Inanc Birol. HLA predictions from the bronchoalveolar lavage fluid and blood samples of eight COVID-19 patients at the pandemic onset. Bioinformatics.
This work was supported by Genome BC and Genome Canada; and the National Institutes of Health.
Learn more about the Birol Lab
Learn more about software development at the GSC
Images created using Biorender.com