News

The genomes of four spruce trees native to North America
Sequencing, assembly and comparative analysis of four spruce giga-genomes completed as part of the SpruceUp project, which aims to enhance the genomic knowledge and accelerate spruce breeding programs across the nation.

Finding Alternatives to Antibiotics in Amphibian and Insect RNA with rAMPage
A bioinformatics pipeline referred to as rAMPage identifies antimicrobial peptides in amphibian and insect RNA sequences.

Transcriptome Analysis Provides Genomic Insight into Emu Fat Metabolism
Understanding gene expression in emu fat tissue could improve production of emu oil and increase its potential health benefits.

Genome polishing: ntEdit+Sealer produces high quality long-read genome assemblies as a genome finishing protocol
The Birol Lab publishes ntEdit+Sealer, an alignment-free genome finishing protocol that corrects sequencing errors and fills gaps to produce highly accurate long-read genome assemblies.
Attentive deep learning model, AMPlify, discovers new antimicrobial peptides (AMPs) from sequencing data
The GSC’s Dr. Inanç Birol Lab publishes AMPlify, an attentive deep learning model that helps predict new, putative antimicrobial peptides (AMPs).
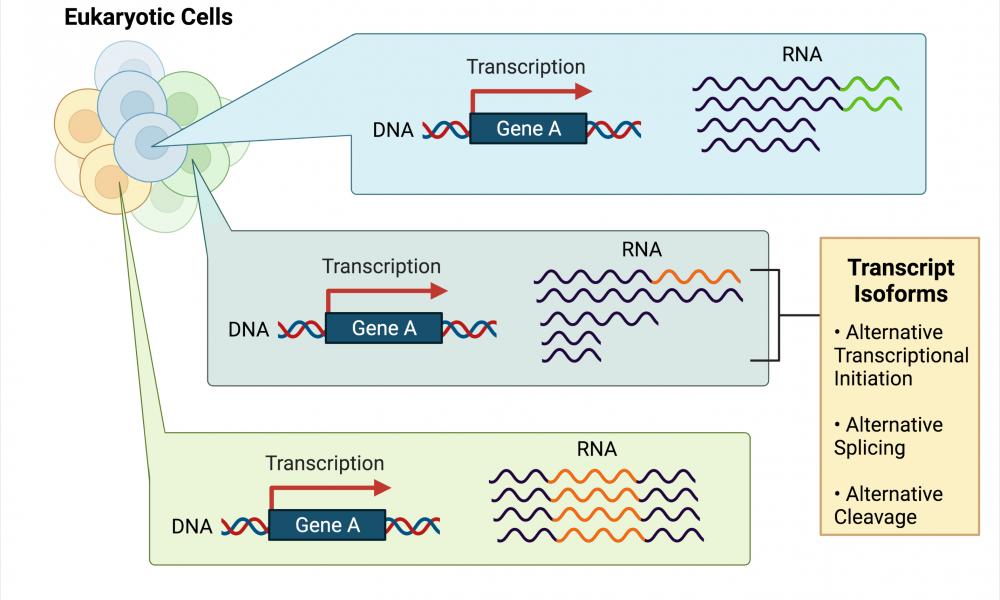
RNA-Scoop: visualizing transcripts from single-cell transcriptomes
The GSC’s Dr. Inanç Birol Lab publishes RNA-Scoop, an interactive tool that allows users to visualize differential transcript expression from single-cell transcriptome datasets.

Suturing draft genomes: LongStitch software helps to mend long read sequences
GSC Researchers from the Birol Lab have developed LongStitch, a scalable pipeline for correcting and assembling draft genomes using long reads.
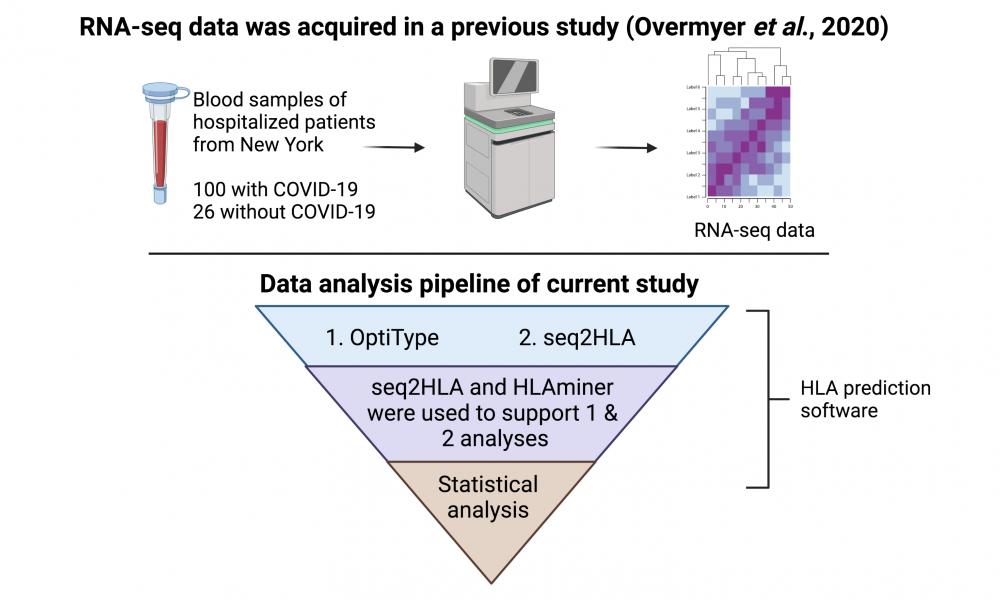
Associating host immune genes with COVID-19 status and disease severity
Using COVID-19 New York patient metatranscriptomics, GSC researchers René Warren and Dr. Inanç Birol, find association between certain HLA alleles and COVID-19 disease severity

Filling in the gaps: GapPredict can complement repertoire of tools used to resolve missing DNA sequences in genome assemblies
GapPredict, the latest bioinformatics innovation from the GSC’s Birol Lab, uses deep learning to predict missing DNA bases in the gaps of [short-read] genome assemblies