The discovery of antibiotics was a pivotal moment for humanity in the battle against human pathogens and disease-causing agents; however, the conflict continues as some pathogens evolved to evade antibiotics. Antimicrobial peptides (AMPs), naturally occurring and known alternatives to conventional antibiotics, represent another mechanism of protection against human pathogens. Researchers from Dr. Inanç Birol’s Lab at the GSC developed an attentive deep learning model called AMPlify that can predict new potential AMPs from sequencing data, effectively shortening the amount of time and resources required to discover AMPs.
Conventional antibiotics versus AMPs
Antibiotic drugs have been relied upon as a medical treatment for certain human pathogens and disease-causing microorganisms. However, over time and due to overuse of antibiotic drugs, some pathogens have evolved resistance against most available antibiotics; such pathogens are called “superbugs”.
In order to fight against this threat, new treatment options are being explored including the likes of antimicrobial peptides (AMPs)—short chains of amino acids (together called peptides) produced naturally by host organisms as part of their innate immune response. Several AMPs have demonstrated antimicrobial activity against bacterial, fungal, and viral pathogens, and shown promise as a non-conventional alternative to conventional antibiotics.
The mechanism of action of AMPs differs from conventional antibiotics as AMPs act more quickly, do not induce resistance in pathogens to the same extent, and can act more broadly by either directly targeting the microorganism (e.g., causing it to rupture) or by enhancing the host immune response. Yet, in spite of these advantages, some pathogens do still develop resistance if exposed to AMPs for extended periods of time.
How to find the most effective AMPs
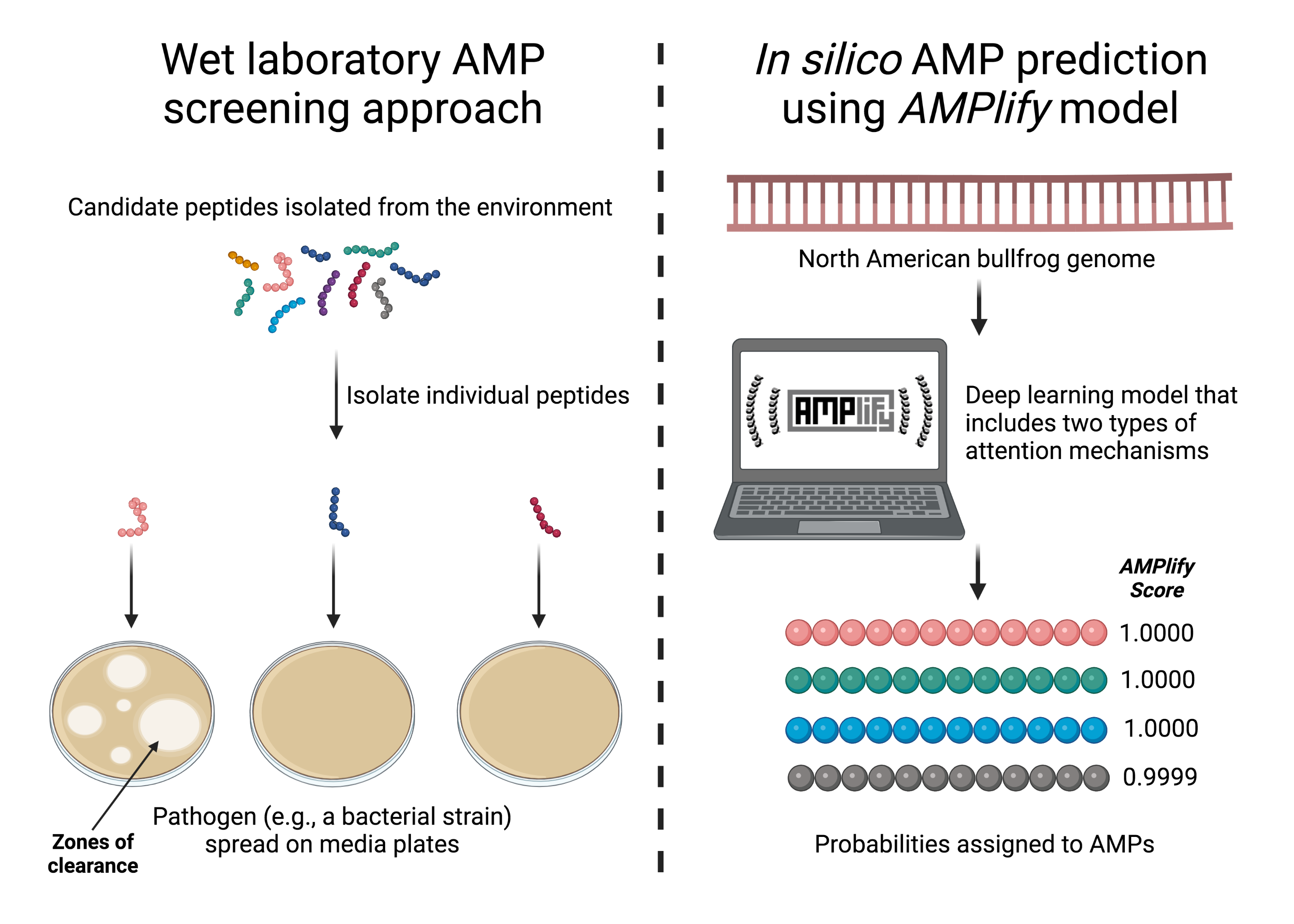
To counteract the continually evolving resistance of such pathogens, fast and accurate methods to discover effective AMPs must be developed. Normally, to discover new AMPs that are effective against human pathogens, large-scale screens are conducted in laboratories. These screens often involve a "soup" of peptides gathered from the environment, where one or more of the peptides in the mixture is thought to have antimicrobial properties.
Next, through laborious searches, scientists try to isolate and characterize the effective peptide. If a specific peptide exhibits antimicrobial activity against a plated pathogen, zones of clearance will be visible on the media plate. Unfortunately, large-scale screening in a laboratory setting is costly, time-consuming and labor-intensive.
Deep neural networks with attention mechanisms
Recently, in silico AMP prediction approaches are being used to aide in the discovery process. Numerous computational models have been developed and each has its share of strengths and weaknesses.
For example, deep learning (a subset of machine learning) is used in many prediction models as these methods can automatically learn high-level features and outperform conventional bioinformatics methods. More recently, deep neural networks with “attention mechanisms”, which resemble how a human brain is capable of prioritizing sections of information when processing text or visual input, are gaining interest for use in bioinformatics deep learning models. In terms of sequence analysis, specific weights are assigned to different positions in a sequence; attention mechanisms work by assessing the specific weights and either amplifying or reducing the importance of the specific sequence at a given position.
Using deep neural networks with attention mechanisms as the basis for their design, researchers in the Birol Lab developed AMPlify, an attentive deep learning model for AMP prediction. As with all machine learning models, AMPlify is trained to recognize AMPs by working on a set of known AMPs and a list of non-AMP peptide sequences. Importantly, AMPlify outperformed existing machine learning methods at predicting AMPs, and is the first AMP prediction model to incorporate attention mechanisms.
Finding putative AMPs using AMPlify
Published recently in BMC Genomics, AMPlify was tasked to search the AMP-rich North American bullfrog genome for known and unknown AMPs. The researchers were able to use AMPlify to successfully identify previously reported AMPs (thereby validating their model) and also found four brand new AMPs. By conducting further in vitro tests, the researchers verified that the four novel AMPs showed antimicrobial activity against a variety of selected bacterial pathogens.
In the future, further improving the AMP discovery and validation pipelines will be critical for identifying and testing new AMPs, some of which will have the potential to replace conventional antibiotics in the fight against superbugs and other human pathogens.
According to the lead author of the publication, Chenkai Li: “While the AMPlify model is very effective for discovering natural AMPs, it also has great potential as a tool for de novo AMP design or enhancement.”
Acknowledgements:
This study was supported financially by Genome Canada and Genome British Columba, and the National Institutes of Health. Additional support was provided by the Canadian Agricultural Partnership, under the Canada-BC Agri-Innovation Program delivered by the Investment Agriculture Foundation of BC.
Image created with BioRender.com. AMPlify logo was created by René L. Warren and Amirhossein Afshinfard.
Learn more:
Learn more about the Birol Lab and their bioinformatics technology research.
Learn about other bioinformatics tools and software for analyzing sequencing data.
Citation:
Li C, Sutherland D, Hammond SA, Yang C, Taho F, Bergman L, Houston S, Warren RL, Wong T, Hoang LMN, Cameron CE, Helbing CC, Birol I. AMPlify: attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens. BMC Genomics. 2022 Jan 25;23(1):77. doi: 10.1186/s12864-022-08310-4. PMID: 35078402.
*bold font indicates members of the GSC.