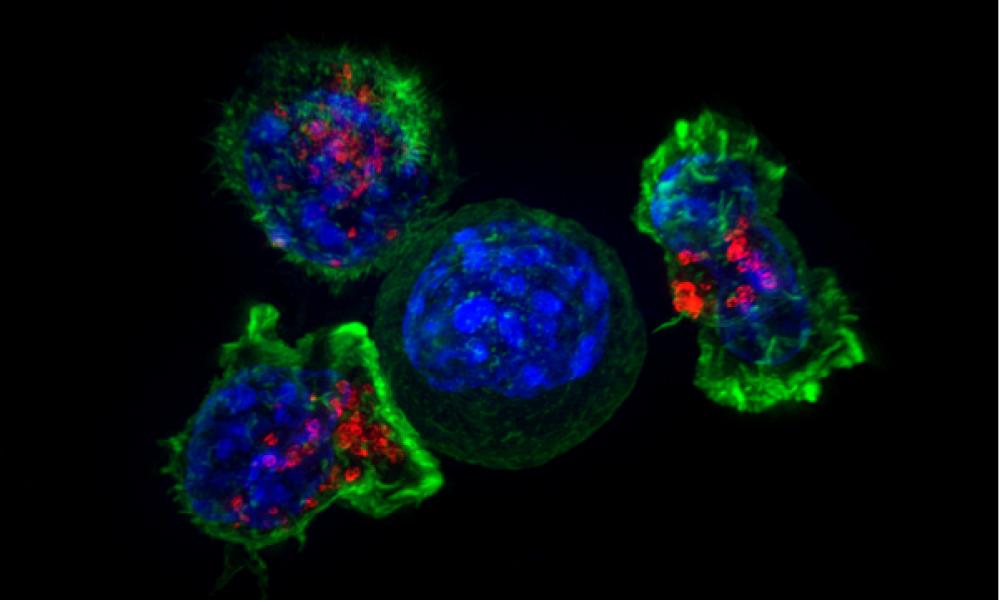
Cancer Immunotherapy is a form of treatment that uses the body’s immune system to identify and attack tumour cells to stop cancer from spreading. Although it can be very effective, the response varies significantly from patient to patient. While concluding that neoantigen counts from noncoding DNA are not informative for predicting patient response to immune checkpoint inhibitors, a study lead by GSC’s Distinguished Scientist Dr. Aly Karsan suggests that characterization of these peptides could significantly expand options for personalized cancer treatment.
Mutations—or DNA changes—are the hallmark of cancer. Since DNA provides instruction on how cells are supposed to function, tumour cells with lots of mutations can behave in ways that make them stand out. This includes production of peptides the cell wouldn’t normally make. Immunotherapy takes advantage of this by helping the body’s immune system recognize and target cells that have these peptides.
Tumour Mutation Burden
To determine which cancers might respond to immunotherapy, scientists measure tumour mutation burden (TMB). The idea behind this is simple: the more DNA mutations there are in a tumour cell, the more likely it is to produce new peptides that could be targeted by the immune system.
The benchmark for measuring TMB is whole exome sequencing, which evaluates the regions of the genome that contains instructions on how to make proteins, also known as coding regions. Noncoding regions—which account for up to 98% of the genome—are typically not included in TMB analysis.
Tumour Neoantigen Count
Peptides produced by tumour cells are called neoantigens. They activate the immune system by binding to the major histocompatibility complex.
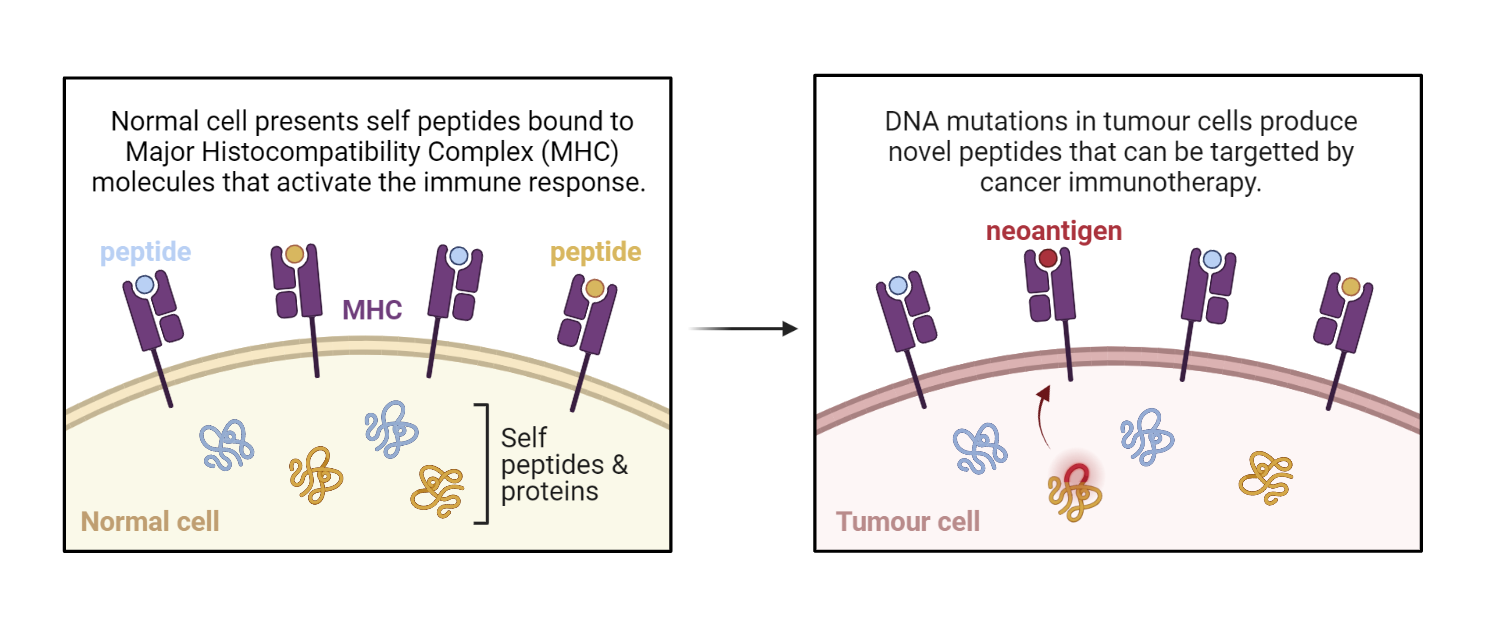
Since high TMB doesn’t guarantee production of new peptides, several tools have been developed to predict neoantigens from DNA-sequencing data. Like TMB, most of these tools are limited to coding regions of the genome.
Comparing TNC to TMB
A study from the Karsan lab used whole genome and transcriptome sequencing to compare TMB and TNC in 574 advanced cancer tumours. Using clinical data from 80 patients who received immunotherapy, the team further compared TMB and TNC as a predictor of response to treatment.
Patients with high TNC in the whole genome and coding regions had a significantly lower chance of disease progression following immunotherapy. This result was similar to the correlation between high TMB and positive patient response to treatment.
In contrast, neoantigens derived from the noncoding regions using transcriptomic data were not informative for predicting patient response. Based on this result, the study authors conclude that assessing neoantigens in the noncoding space does not help determine which patients will respond to immunotherapy over standard TMB analysis.
Implications and Future Directions
“We've developed a pipeline to predict neoantigens arising from the aberrant expression of sequences in the intergenic region or from retained introns that are normally spliced out of the mature transcript,” says Tammy Lau, first author of the publication and former research programmer in the Karsan lab. Intragenic regions refer to noncoding DNA between genes, whereas introns are noncoding DNA sequences located within protein-coding genes.
“Although having more neoantigens predicted from these sources doesn't inform whether a patient will respond to immunotherapy in the form of immune checkpoint inhibitors, we can still try to validate some of these predicted neoantigens experimentally using a proteomics screen.”
According to Tammy, this could drastically expand the search for personalized treatment options beyond immune checkpoint inhibitors, since the majority of the genome is noncoding. “Others have already shown that the expression of introns can produce neoantigenic peptides, but little work has been done in exploration of the intergenic region of the genome."
Acknowledgements:
The authors thank the patients and families, the Personalized OncoGenomic team, Canada’s Michael Smith Genome Sciences Center technical platforms, and the generous support of the BC Cancer Foundation and their donors.
This work was supported by a grant to AK from the Collaborative Health Research Projects through the Canadian Institutes of Health Research (CPG-146474) and the Natural Sciences and Engineering Research Council of Canada (CHRP 493628-16), and Genome British Columbia (#X01CBP). AK is supported by the John Auston BC Cancer Foundation Clinician-Scientist Award.
Killer T Cells Surrounding a Cancer Cell Banner Image Credit: National Institutes of Health (Alex Ritter, Jennifer Lippincott Schwartz, and Gillian Griffiths).
Neoantigen Image created with biorender.com
Learn More:
Learn more about cancer immunotherapy.
Learn more about the Karsan lab and GSC research on immune checkpoint inhibitors.
Citation:
Tammy T.Y. Lau, Zahra Jalali Sefid Dashti, Emma Titmuss, Alexandra Pender, James T. Topham, Joshua Bridgers, Jonathan M. Loree, Xiaolan Feng, Erin D. Pleasance, Daniel J. Renouf, Kasmintan A. Schrader, Sophie Sun, Cheryl Ho, Marco Marra, Janessa Laskin, Aly Karsan. The neoantigen landscape of the coding and noncoding cancer genome space. Journal of Molecular Diagnostics.
*bold font indicates members of the GSC.