Mantle cell lymphoma (MCL) is a rare and incurable form of B cell non-Hodgkin lymphoma. To develop effective cancer therapies for the disease, a better understanding of how MCL cells develop and persist is essential.
In a new study published in the journal Blood, led by GSC Senior Scientist Dr. Ryan Morin, researchers used exome, genome and transcriptome sequencing to characterize MCL tumours and identified a novel feature of MCL biology.
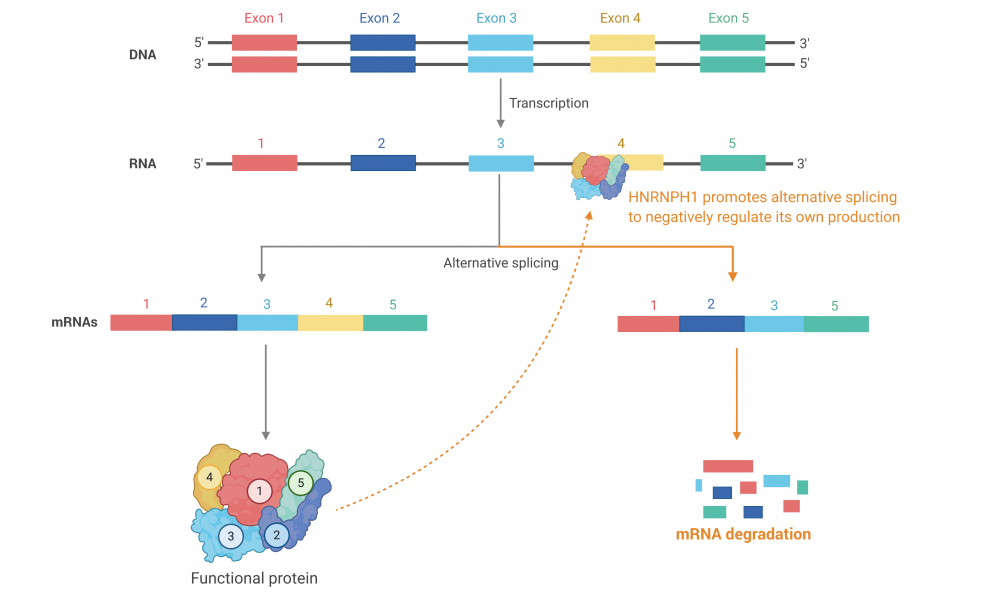
Their analyses revealed that MCL tumours harbored recurrent non-coding mutations in the gene encoding HNRNPH1, a protein that binds to mRNA and regulates alternative splicing events.
In a normal cell, HNRNPH1 regulates its own production. It binds to HNRNPH1 mRNA resulting in an alternative splicing pattern that excludes exon 4, leading to degradation of the shortened mRNA isoform through a process called nonsense-mediated decay. This regulatory mechanism limits the amount of functional HNRNPH1 protein in a cell at any one time, maintaining alternative mRNA splicing events at an optimal level.
The non-coding mutations detected in HNRNPH1 were found to disrupt this intricate regulatory mechanism, leading to over-production of functional HNRNPH1. Higher levels of HNRNPH1 were associated with inferior disease outcomes, implying that disrupted mRNA processing may play an important role in MCL.
"Finding recurrent mutations in HNRNPH1 and in other RNA-processing genes, such as DAZAP1 and EWSR1, highlights the role that RNA-processing plays in MCL. RNA-processing proteins affect the abundance and functioning of many other proteins by regulating how, when and if mRNA are generated and small changes to these regulatory proteins via substitutions can have large consequences within the tumour cell," says Prasath Pararajalingam, graduate student in Dr. Morin's laboratory and, together with Dr. Krysta Coyle, first author on the study.
In addition to confirming previously identified TP53 and NOTCH1 mutations as indicators of prognosis, this study revealed the association between non-coding HNRNPH1 mutations and disease outcomes in MCL patients. Further, the study uncovered a novel aspect of MCL biology, improving our understanding of the disease and providing a starting point for the development of novel treatment strategies.
Prasath Pararajalingam, Krysta M Coyle, Sarah Arthur, Nicole Thomas, Miguel Alcaide, Barbara Meissner, Merrill Boyle, Quratulain Qureshi, Bruno M Grande, Christopher Rushton, Graham W Slack, Andrew Mungall, Constantine Tam, Rishu Agarwal, Sarah-Jane Dawson, Georg Lenz, Sriram Balasubramanian, Randy D Gascoyne, Christian Steidl, Joseph Connors, Diego Villa, Timothy E Audas, Marco A Marra, Nathalie A Johnson, David W Scott, Ryan D Morin. 2020. Coding and non-coding drivers of mantle cell lymphoma identified through exome and genome sequencing. Blood. 136(5):572-584.
Learn more about research in Dr. Ryan Morin's lab
Learn more about research at the GSC