Large-scale genome sequencing initiatives frequently rely on datasets provided by different sequencing centres, all of which may employ different laboratory protocols, instruments and analytical methods that may introduce disparities in the data produced. When combining datasets generated at different institutions, how do researchers ensure consistency between datasets?
In early 2019, Dr. Andy Mungall, Biospecimen and Library Cores Group Leader at the GSC and former Chair of the Canada Genomics Enterprise (CGEn) Technical Experts Committee, co-supervised an experiment to address this disparity. Consequently, this has also primed the three centres that comprise CGEn to embark on a national population sequencing initiative aimed at understanding host factors impacting COVID-19 disease outcomes.
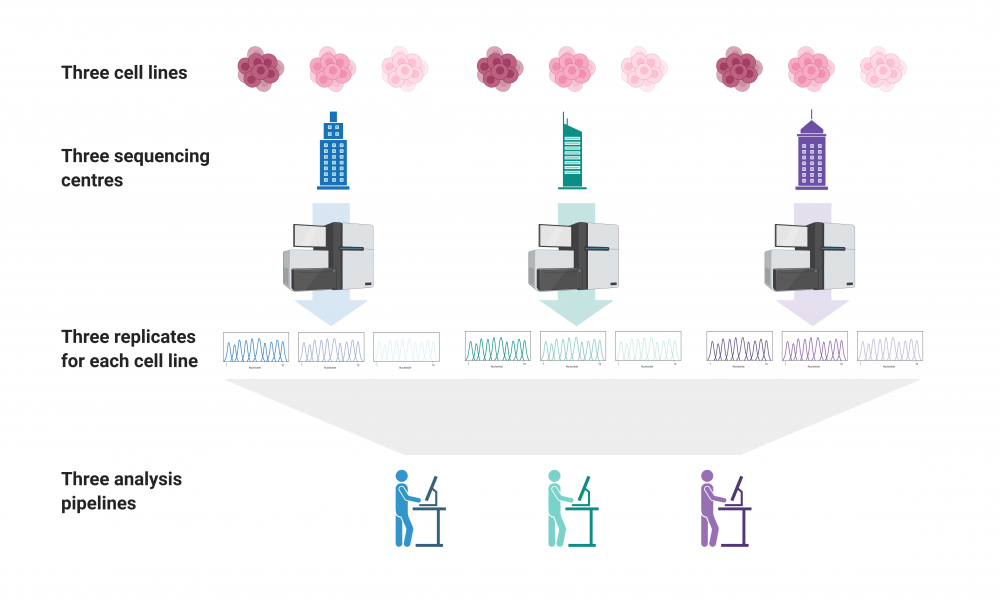
In the study, published in the journal Frontiers in Genetics, researchers from CGEn’s three nodes—the GSC, The Centre for Applied Genomics at The Hospital for Sick Children in Toronto and the McGill Genome Centre in Montreal—compared sequencing data produced and analyzed at each of the three centres.
To facilitate their evaluation, each node sequenced the genomes of three standard reference cell lines from the Genome in a Bottle (GIAB) consortium. The consortium enables sequencing centers to routinely assess the precision and sensitivity of single nucleotide variants (SNVs) and insertion and deletions (indels) detected in their analyses by sequencing GIAB reference samples.
“The idea of this experiment was to see how comparable data independently generated at the three sites were to one another, and whether CGEn might lend itself to national-scale population sequencing,” says Dr. Mungall.
The team used three replicates of three different DNA samples from the GIAB consortium, sequenced them at the three sequencing centres and analyzed the data using three different bioinformatics pipelines, resulting in a total of 81 different datasets for comparison.
Their results demonstrated that datasets produced at each of the three centres were highly similar, and that the most significant differences were the result of the analytical pipelines used rather than laboratory methods and instrumentation.
“We were very pleased to find that despite using different laboratory methods, the sequencing data produced at the three centres were very consistent. In fact, they were just as consistent as replicates sequenced at one centre. That was very reassuring,” says Dr. Mungall. “Data analysis is dependent on the experiment being conducted; all the three centres specialize in different types of analyses, so we were not surprised to find some disparities arising due to different analytical pipelines. On the whole, however, data production is very similar across Canada’s three high-throughput genome sequencing centres.”
CanCOGeN
As part of the Canadian COVID Genomics Network (CanCOGeN)—a Genome Canada initiative that will generate accessible and usable genomics data to inform our COVID-19 public health decisions—CGEn will sequence 10,000 Canadians that have tested positive for the virus.
“This study gives us confidence that we are primed and ready for national-scale population sequencing projects like the CanCOGeN HostSeq initiative,” says Dr. Mungall. “Sequencing of the 10,000 human genomes will be distributed across CGEn’s three centres, and we now know we need to pay particular attention to how those data are analyzed. To do this, packaged and distributable analysis pipelines will be shared between the centres, ensuring that all of the datasets are processed through an identical analytical pipeline.”
More than 500,000 human genomes have been sequenced and deposited in public databases to date. These data have provided the foundation for enhanced understanding of the genetic basis of human health and disease and are increasingly being used in clinical genetics settings as a biological reference for national precision medicine initiatives. As more large-scale, multi-site sequencing projects are launched, researchers will need to evaluate datasets produced at different sequencing centres to ensure high confidence in the pan-geographic reproducibility of results.
This study provides a framework for evaluating consistency in data generated in national and international distributed genome sequencing projects. Further, by publishing the study in Frontiers in Genetics, the manuscript and datasets are publicly available. This study focused on SNVs and indels, but researchers are able to dive deeper into the data to analyze structural variants, copy number variants or other genomic features of interest to see what differences may be expected from a distributed network.
Learn more about the Canada Genomics Enterprise
Learn more about the McGill Genome Centre
Learn more about The Centre for Applied Genomics
Richard D. Corbett, Robert Eveleigh, Joe Whitney, Namrata Barai, Mathieu Bourgey, Eric Chuah, Joanne Johnson, Richard A. Moore, Neda Moradin, Karen L. Mungall, Sergio Pereira, Miriam S. Reuter, Bhooma Thiruvahindrapuram, Richard F. Wintle, Jiannis Ragoussis, Lisa J. Strug, Jo-Anne Herbrick, Naveed Aziz, Steven J. M. Jones, Mark Lathrop, Stephen W. Scherer, Alfredo Staffa and Andrew J. Mungall. A Distributed Whole Genome Sequencing Benchmark Study. Frontiers in Genetics. 11:612515.
CGEn is a national sequencing facility supported by the Canada Foundation for Innovation’s Major Science Initiatives. Leveraged co-funding supporting CGEn and the experiments was provided by Genome Canada through Genome BC, Ontario Genomics, and Génome Quebec, and by the University of Toronto McLaughlin Centre.