Studying human diseases like cancer is challenging for a variety of reasons; one important consideration is the accuracy of the model systems used to study cancers in the laboratory. Researchers often rely on animal models, human cell lines, or both to create “cancer-like” conditions. In recent years, a newer type of model system known as “patient-derived tumour explants” has emerged as a promising tool to study cancers and test treatments. A recent study by GSC scientists explores the extent to which explants mimic brain tumours.
Three-dimensional patient-derived models: an alternative way to study diseases
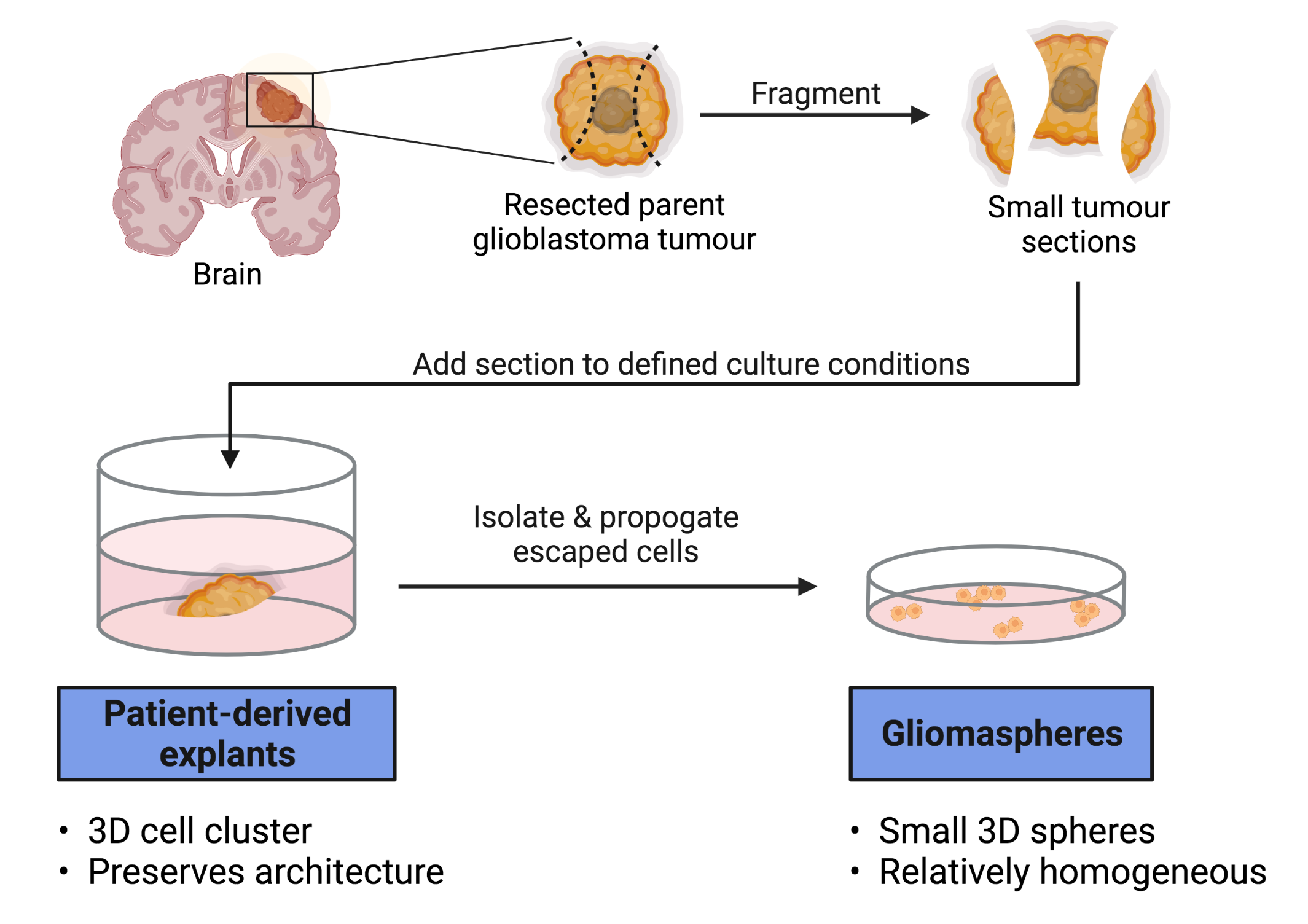
Patient-derived explants (PDEs, also sometimes called “tumour organoids”)—a type of model system (ex vivo) used to study diseases—are made by implanting a small piece of the tumour under study into a gel bead and allowing the tumour to grow out into the gel, thereby keeping the 3D structure of the tumour intact.
By preserving the structure and the 3D interactions between cells, important tumour features are retained. This is in contrast to other model systems where these features can be lost (e.g., human cell lines that are grown in 2D). Although PDE models are promising, the extent to which they capture the features of individual patient tumours is important to investigate in order to evaluate their potential for research and clinical studies.
Tumour heterogeneity
Tumours are usually made up of non-identical cells, thus making the tumour “heterogeneous”. The cells that make up a single tumour can have different genetic and functional characteristics, including healthy “non-cancerous” cells that infiltrate the tumour from the surrounding normal tissue. The observation that cells within a single tumour are not identical to each other is broadly referred to as ‘intra-tumour heterogeneity’. Furthermore, even tumours of the same cancer type are often different from each other at the genetic and cellular levels—this is known as “inter-tumour heterogeneity”.
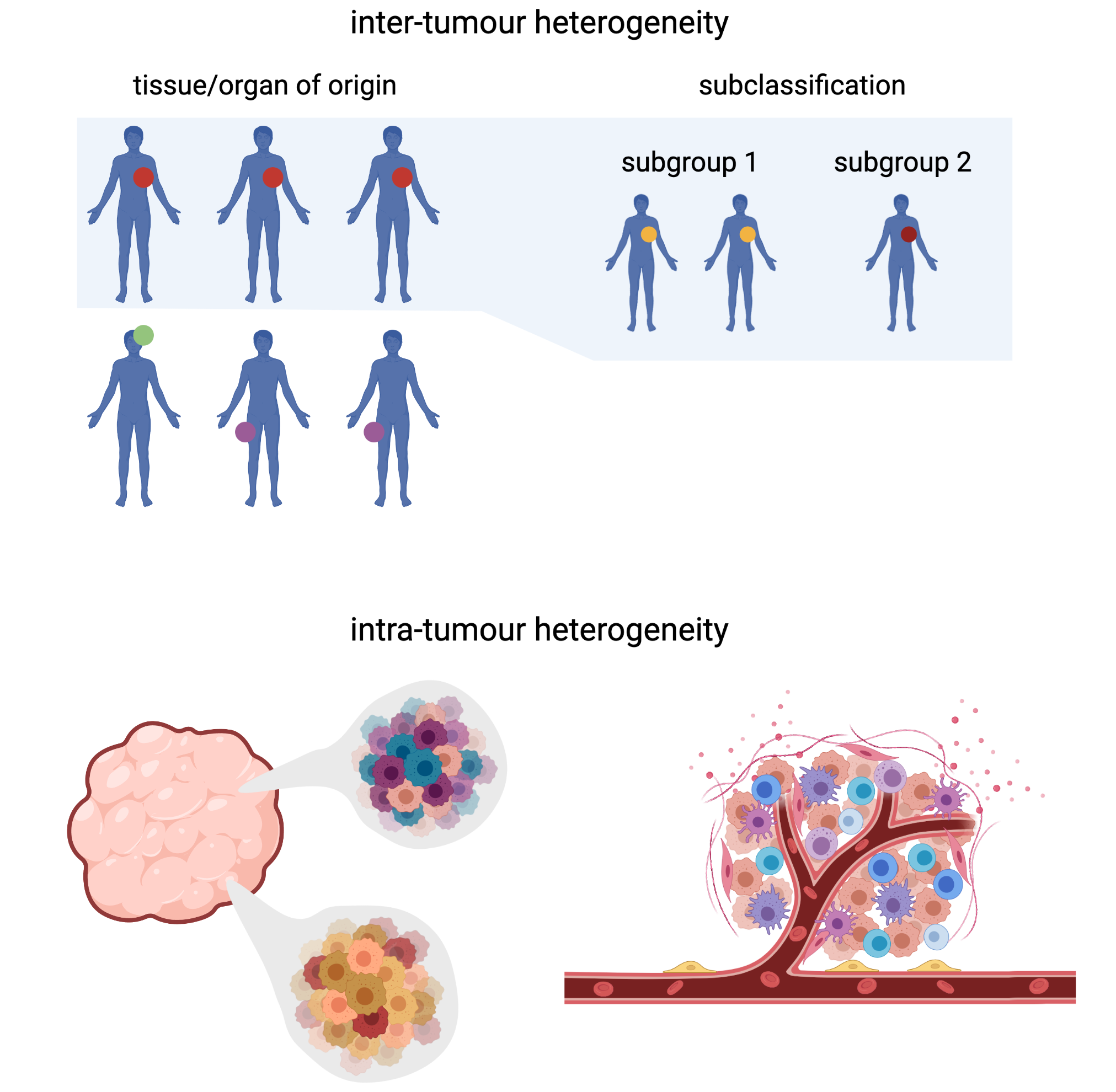
High levels of tumour heterogeneity, especially the intra-tumour kind, tend to be linked with more aggressive disease; therefore, it is important for scientists to be able to model tumour heterogeneity accurately in the laboratory to better understand and manage disease.
Single-cell sequencing
Sequencing technologies developed over the past few decades have allowed us to understand the molecular (e.g., genetic) characteristics of tumours. However, most sequencing technologies rely on average measurements over a population of cells, and thus cannot capture the full extent of tumour heterogeneity.
More recently, new sequencing technologies have emerged that allow us to obtain measurements from individual cells—so-called “single-cell sequencing”. Using single-cell sequencing approaches, scientists have learned a great deal about not only the genetic features of cells, but also their observable traits (i.e., their “phenotype”); for example, their patterns of RNA expression, or “transcriptional” characteristics.
Glioblastomas are heterogeneous tumours in need of better models
Glioblastomas (GBMs)—the most common type of malignant brain cancer—are a prime example of cancers that display particularly high inter- and intra-tumour heterogeneity. GBM tumours are also very difficult to treat; thus, patients have few therapeutic options. Developing accurate models of this tumour that are accessible in the laboratory is critical to developing a better understanding of this cancer and to translating that knowledge to better therapies.
While GBM-derived explants have recently been developed, whether they accurately mimic the patient-specific heterogeneity of GBM tumours remains unclear. From the lab of Distinguished Scientist and GSC Director Dr. Marco Marra, researchers sought to answer this question by investigating the heterogeneity of GBM tumours and models derived from them, including PDEs.
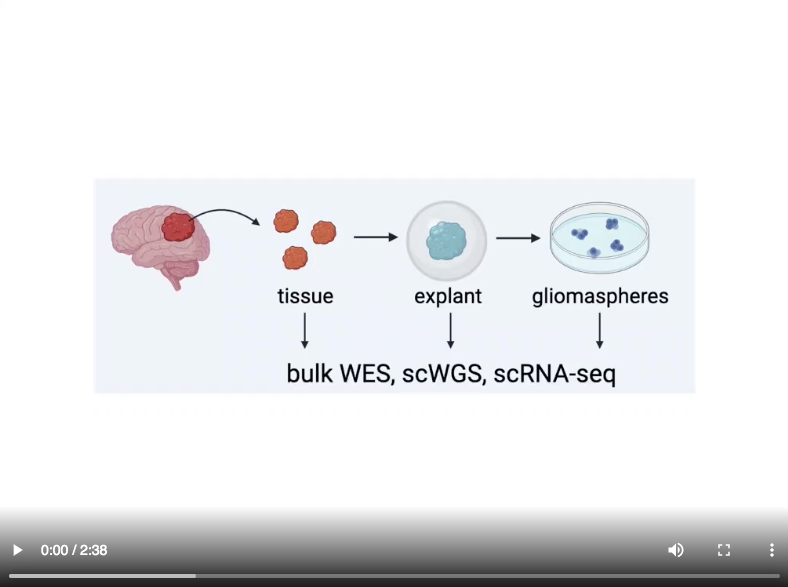
In an article recently published in Cancer Cell, the Marra Lab used sequencing approaches—including single-cell DNA sequencing (scDNA-seq) and RNA sequencing (scRNA-seq)—to analyze the genetic and transcriptional heterogeneity of GBMs, PDEs, and more traditional GBM cell lines (propagated as so-called “gliomaspheres”) derived from the same tumours.
“Single-cell sequencing approaches have transformed our understanding of GBM biology and especially heterogeneity,” says Dr. Véronique LeBlanc, postdoctoral fellow in the Marra Lab and lead author of the study. “By using such methods to profile matched tumours and model systems, we were able to generate a large-scale dataset that we believe will provide a valuable resource for the GBM research community to continue investigating both GBM biology and the relative utility of different model systems.”
The researchers were able to show that GBM PDEs are generally genetically similar to their parent tumours. They also found that GBM PDEs exhibited similar levels of transcriptional heterogeneity as their parent tissues, whereas cell lines tended to be composed of cells more similar to one another (i.e., more “homogeneous”). Finally, the authors also showed that PDEs retained this heterogeneity over multiple generations, indicating that they could potentially be used to test novel therapeutics.
These results imply that PDEs may present a more representative model with which to study heterogeneity in GBM compared to cell lines. They also highlight the importance of using a single-cell approach to characterize model cancer systems before using them to conduct experiments, in order to verify that they accurately represent the cancer under study. In addition, these results offer a foundation for further investigation into the underlying biology of GBM.
“We hope that, by underscoring the importance of deep genomic profiling of model systems to ensure they are fit for purpose, our study will prompt an increase in the use of similar approaches to ensure the validity of model systems across cancers,” says Dr. LeBlanc.
Acknowledgements:
This study was funded by the Terry Fox Research Institute and the Canadian Institutes of Health Research.
PDE image created with BioRender.com and adapted from Figure 1A of the Cancer Cell publication by LeBlanc et al. (2022). Thank you to Drs. Jennifer Chan and Bo Young Ahn for allowing us to use their immunofluorescence image as the title figure for this story. Thank you to Dr. Véronique LeBlanc for allowing us to use her tumour heterogeneity figure (created using BioRender).
Learn more:
Learn more about the Marra Lab and their other glioma-related projects at the GSC.
Learn more about glioblastoma heterogeneity.
Citation:
LeBlanc VG, Trinh DL, Aslanpour S, Hughes M, Livingstone D, Jin D, Ahn BY, Blough MD, Cairncross JG, Chan JA, Kelly JJ, Marra MA. Single-cell landscapes of primary glioblastomas and matched explants and cell lines reveal variable retention of inter- and intra-tumor heterogeneity. Cancer Cell. 2022 Mar 14:40:S1535-6108(22)00068-X. doi:10.1016/j.ccell.2022.02.016. Epub ahead of print. PMID: 35303420.
*bold font indicates members of the GSC.