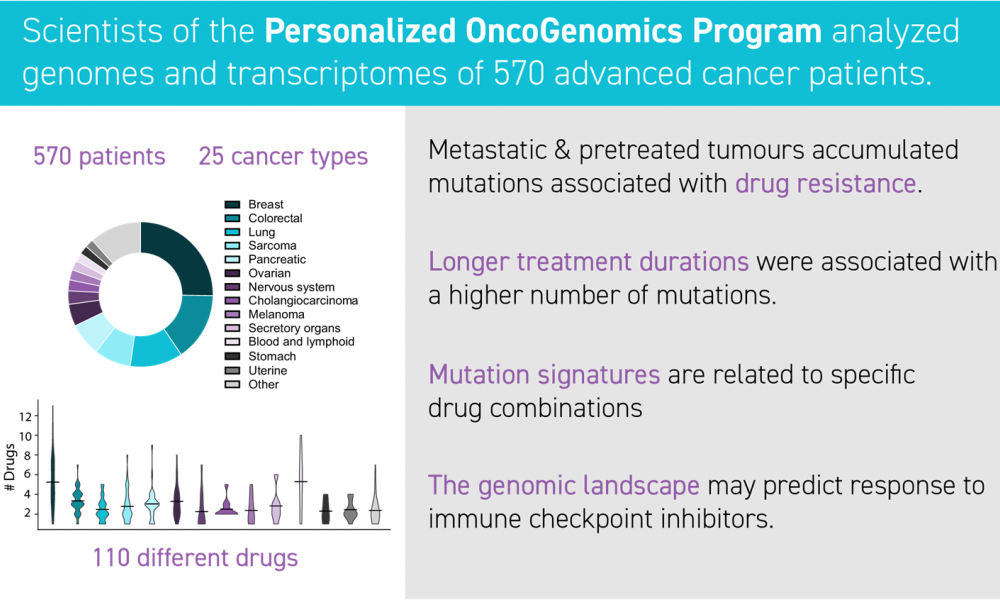
A comprehensive analysis of 570 advanced cancer patients has revealed the genetic changes that occur following chemotherapy, providing scientists and clinicians with valuable insight into advanced cancers and drug resistance.
Despite the significant progress since the advent of next generation sequencing technologies in understanding the genetic alterations that lead to cancer formation, relatively little is known about how cancers progress, spread throughout the body and become resistant to chemotherapy. But, as the majority of cancer deaths occur following treatment failure, analysis of the genetic alterations that contribute to drug resistance in advanced cancer patients is critical.
In a study focused entirely on DNA mutations that occur in advanced-stage cancers, researchers thoroughly scrutinized genomic, transcriptomic and clinical data to shed light on the genetic alterations that occur following cancer treatment and spread.
“This study is significant because it presents the most intensively analyzed cohort in B.C. of heavily-treated cancers, and tries to relate these data to cancer treatment, both duration and type,” said Dr. Marco Marra, Director of Canada’s Michael Smith Genome Sciences Centre (GSC) at BC Cancer and lead researcher of the study.
Personalized OncoGenomics
The study, published in Nature Cancer, is part of the Personalized OncoGenomics (POG) program at BC Cancer led by Dr. Marra and medical oncologist Dr. Janessa Laskin. Together, they lead an interdisciplinary team of scientists, oncologists, pathologists, medical geneticists, interventional radiologists and surgeons to revolutionize our understanding of the use of genomics for cancer treatment planning.
The program, which has enrolled more than 1,000 individuals to date, conducts intensive analyses of genomic, transcriptomic and clinical data to influence cancer treatment planning for individual patients while also producing an extensive dataset that can be interrogated by researchers around the world.
“Not only are patients being treated as they come in, providing them with treatment options which overall improves survival of POG patients,” said Dr. Erin Pleasance, staff scientist at the GSC and author on the study. “But we are also amassing a huge amount of genomic and clinical data on advanced cancers. This allows us to do retrospective analyses to gain critical insight into advanced cancers.”
The scars of cancer treatment
Many cancer patients initially respond to chemotherapy, but often, their cancers return with devastating outcomes. Within each tumour are populations of cancer cells, each with a different set of genetic mutations. Chemotherapy that is effective often eliminates only a subset of sensitive cancer cells while selecting for the continued growth of those that are resistant to that same treatment, leading to cancer progression and ultimately treatment failure.
This in-depth analysis of the first 570 patients to be enrolled in the POG program, aims to gain insight into this mechanism of cancer drug resistance and spread. The cohort was comprised of a diverse group of patients representing 25 different cancer types from 18 anatomical locations, enabling researchers to look broadly at genetic alterations that occur across many different cancers.
“Most cancer analyses concern themselves with division of cases into histologically similar tumour types,” said Dr. Marra. “This study attempts to go beyond that and address issues that cut across malignancies.”
Importantly, more than 80 per cent of patients had received at least one of the 110 different cancer medications used in this cohort prior to enrollment; more than 75 per cent of tumour samples were taken after the cancer had spread throughout the body.
The researchers found evidence of prior cancer treatment written within the patients’ DNA. Their analyses revealed that tumours exposed to prior treatment increased the number of mutations, some of which are associated with resistance to chemotherapies.
“The mutations we observed were consistent with patterns that have been described in initial, untreated tumours,” says Dr. Laura Williamson, staff scientist at the GSC and author on the study. “But there were also huge additional changes in these samples that relate to therapy across different tumour types.”
Chemotherapy treatment was associated with an increased number of mutations and the longer patients were treated, the more mutations they had. These findings provide important insight into genetic alterations that allow cancer cells to thwart treatment.
“Patients who were treated with these drugs prior to biopsy were found to have an increased mutation burden, some of which are likely associated with resistance,” said Emma Titmuss, research programmer at the GSC and author on the study.
Predicting patient outcomes
The authors also identified patterns of genomic alterations that are associated with the immune system—our body’s natural defense against diseases including cancers. These patterns (or gene expression profiles) correlated with patient survival and response to a type of chemotherapy called immune checkpoint inhibitors. These findings may enable clinicians to predict which patients will respond well these therapies and which patients will not.
“By looking at the genome landscape we can have some predictive power to say whether patients will respond to immune checkpoint inhibitors based on the pattern of alterations that we see,” said Dr. Pleasance.
Enabling future research
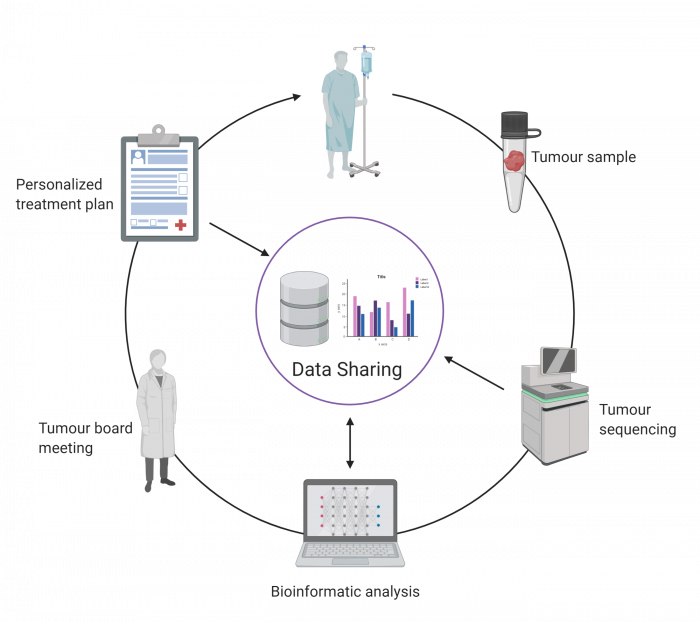
Patients enrolled in the POG program consent to having their genomic data anonymously shared with the broader scientific community, each patient thereby contributing to the advancement of knowledge for the benefit of patients around the world. Sharing of these data is vitally important for cancer researchers to discover subtle genetic patterns that would be impossible to detect in one individual, revealing themselves only when hundreds of datasets are combined and analyzed as a whole.
“In addition to the analyses themselves, we provide the data to the research community so that they can look at particular drugs they are interested in, and explore how their drug behaves in our data,” said Dr. Pleasance.
“The availability of data from heavily-treated individuals is going to stimulate analyses in many different ways,” said Dr. Marra. “I think people will be trying to mine our data for signals that they can leverage across the full spectrum of cancer research. I do believe that the initiative has shown that, in terms of data sharing, you can collect these kinds of data from patients that are undergoing treatment, and that patients want to share their data. Finding subtle clues from the genome and then using these clues to build hypotheses is what science is all about. I can’t imagine all of the possible ways that people will use our data, but it will be available and it will be used.”
The impact of POG
It has been nearly ten years since the GSC’s landmark publication in Genome Biology demonstrating for the first time the utility of whole genome sequencing for cancer treatment planning, inspiring the development of the POG program and cancer genomics research across the country.
“The POG program has led the way for this country to imagine working on pre-treated and resistant cancers in this particular way and to establish a network of cancer centres for data sharing on uniformly collected datasets,” said Dr. Marra, “It has started something that goes beyond our own scientific discoveries. It has started a revolution.”