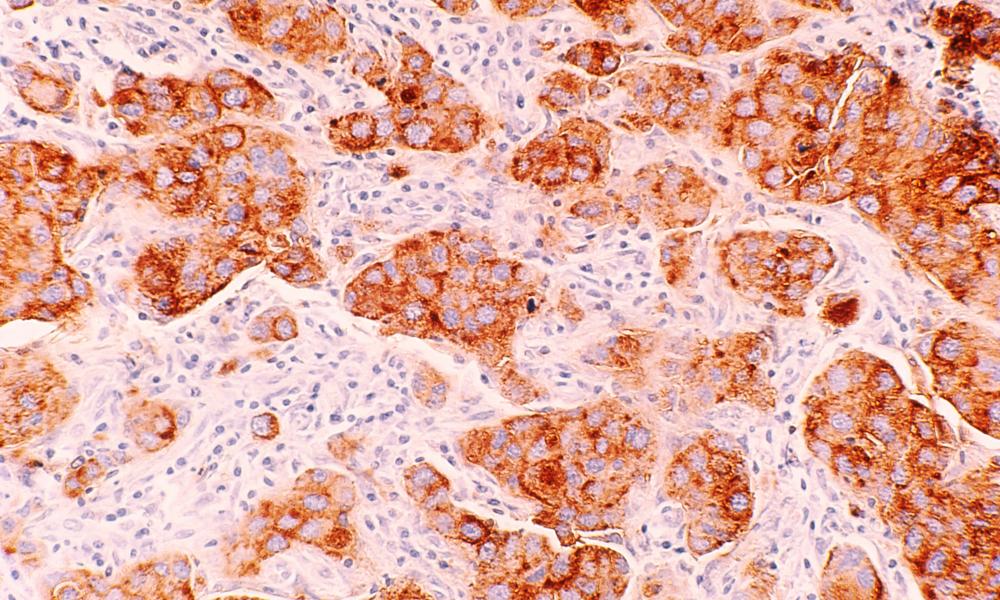
Collectively, breast cancer—also referred to as breast carcinoma—is the most common cancer in women worldwide. Breast cancer is recognized as a family of diseases, each considered a breast cancer subtype, but how these subtypes are defined is still evolving. Dr. Gregg Morin and the GSC Proteomics Platform worked with VGH and BC Cancer collaborators to conduct large-scale proteome-based profiling using formalin-fixed paraffin-embedded (FFPE) tissue specimens and were able to identify novel breast cancer subtypes with distinct clinical outcomes.
Classifying breast cancer subtypes
Breast cancer—recognized as a family of diseases rather than a single disease—can be divided into multiple subtypes, with each tumour showing distinct clinical, pathological, and molecular features.
Breast cancers can be classified into subtypes by profiling genomic properties, including DNA and RNA alterations (e.g., the Prediction Analysis Microarray 50 (PAM50) RNA-based gene signature). Using genomic profiling for the breast cancer classification process has greatly advanced breast cancer diagnosis and improved our ability to predict patient outcomes, including response to treatment.
However, genomic profiling is not always used to decide therapy options, as breast cancers as a group exhibit considerable heterogeneity in their underlying causes and vulnerabilities, thus complicating treatment decisions. Additionally, alterations to DNA/RNA do not always translate into protein-level differences for biomarker or target discovery; this is important to consider as therapies usually target biological changes at the protein level.
Creating a clinically-applicable breast cancer classification system
Researchers from the GSC’s Dr. Gregg Morin Lab, along with their collaborators, Dr. Torsten Nielson (breast cancer pathologist, VGH) and Dr. Stephen Chia (oncologist, BC Cancer), aimed to develop a new classification system for breast cancer with potential clinical applications. For this system, they used standard formalin-fixed paraffin-embedded (FFPE) clinical samples—the most widely available biorepository materials and routinely used for patient diagnosis.
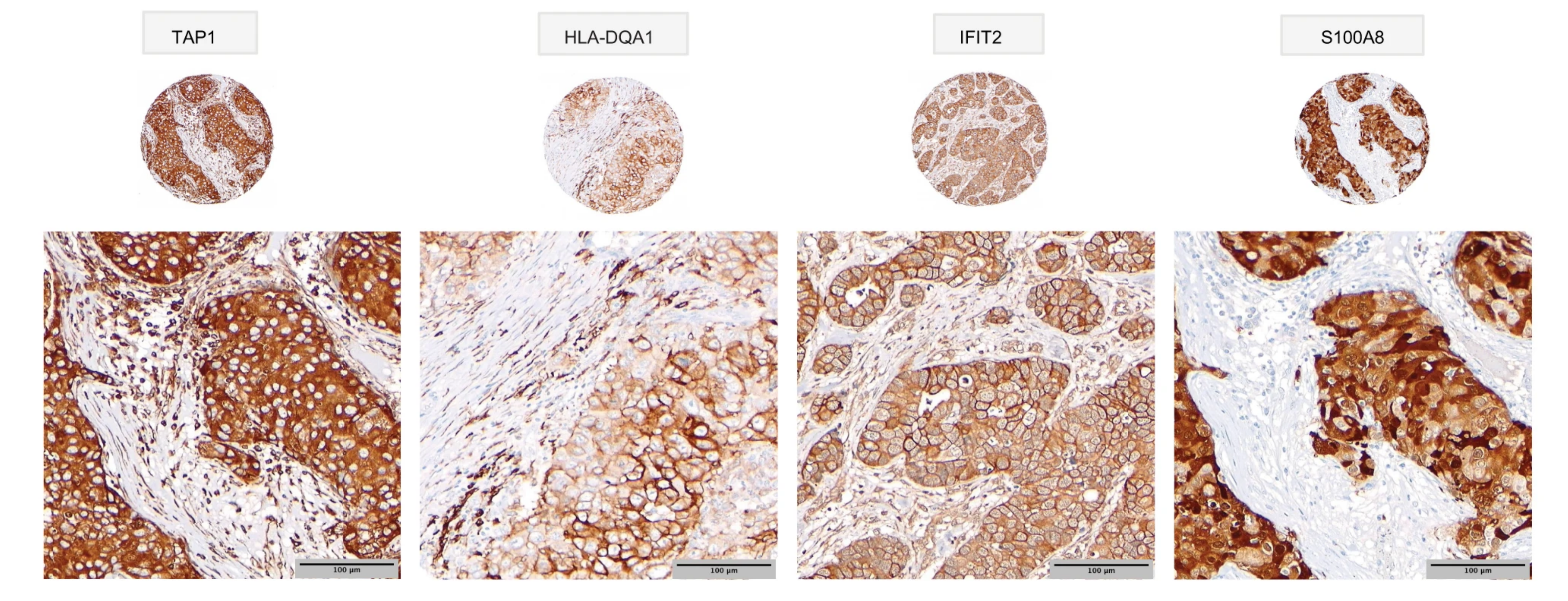
In their study, published in Nature Communications, the researchers identified abnormally expressed proteins in 300 clinical breast cancer FFPE tissue samples, archived since the mid-1980s, thus providing long term survival data. From their analysis, not only were they able to identify groups of specifically expressed proteins, they also noted that these groups coincided with patient subgroups of both poor and good long-term breast cancer survival outcomes.
For example, in triple negative breast cancers (TNBC)—an aggressive breast cancer subtype with generally poor outlook—their data showed that TNBC could be further divided into subgroups with poor survival and, most significantly, a group with highly favourable survival. Based on their findings, the research suggests this “good” prognosis group may benefit from treatments avoiding chemotherapy and its damaging side effects.
Evaluating the clinical impact and future steps
In summary, the researchers characterized distinct proteome groupings, identified potential biomarkers, and matched the identified breast cancer subtypes to detailed clinical outcomes. Notably, the researchers found that specific breast cancer subtypes with distinct survival outcomes could be identified from the FFPE samples using a proteomics-based approach. Their approach can also be applied to other FFPE tumour-banked samples. With this technology, researchers and clinicians can survey large numbers of archived patient tumour samples in other cancer types to identify patient outcome subgroups and associated diagnostic/prognostic biomarkers or new drug candidates.
The research conducted here reveals the heterogeneity in breast cancer and identifies new subgroups of TNBC linked to both poor and highly favourable survival or “good” patient outcomes. In the future, with an award from the Canadian Cancer Society, the researchers aim to develop a clinical assay to identify “good” TNBC patients—those unlikely to benefit from aggressive chemotherapy—and redirect such patients to other, better-suited treatment options.
Acknowledgements:
This study was supported financially by the Canadian Cancer Society.
Learn more:
Learn more about the ongoing research by the Gregg Morin Lab at the GSC.
Learn more about the GSC’s Proteomics Platform, overseen by Dr. Gregg Morin, Head of the Proteomics Platform.
Learn more about the ongoing research by Dr. Torsten Nielson and Dr. Stephen Chia.
Citation:
Asleh K, Negri GL, Spencer Miko SE, Colborne S, Hughes CS, Wang XQ, Gao D, Gilks CB, Chia SKL, Nielsen TO, Morin GB. Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes. Nat Commun. 2022 Feb 16;13(1):896. doi: 10.1038/s41467-022-28524-0. PMID: 35173148; PMCID: PMC8850446.
*bold font indicates members of the GSC.