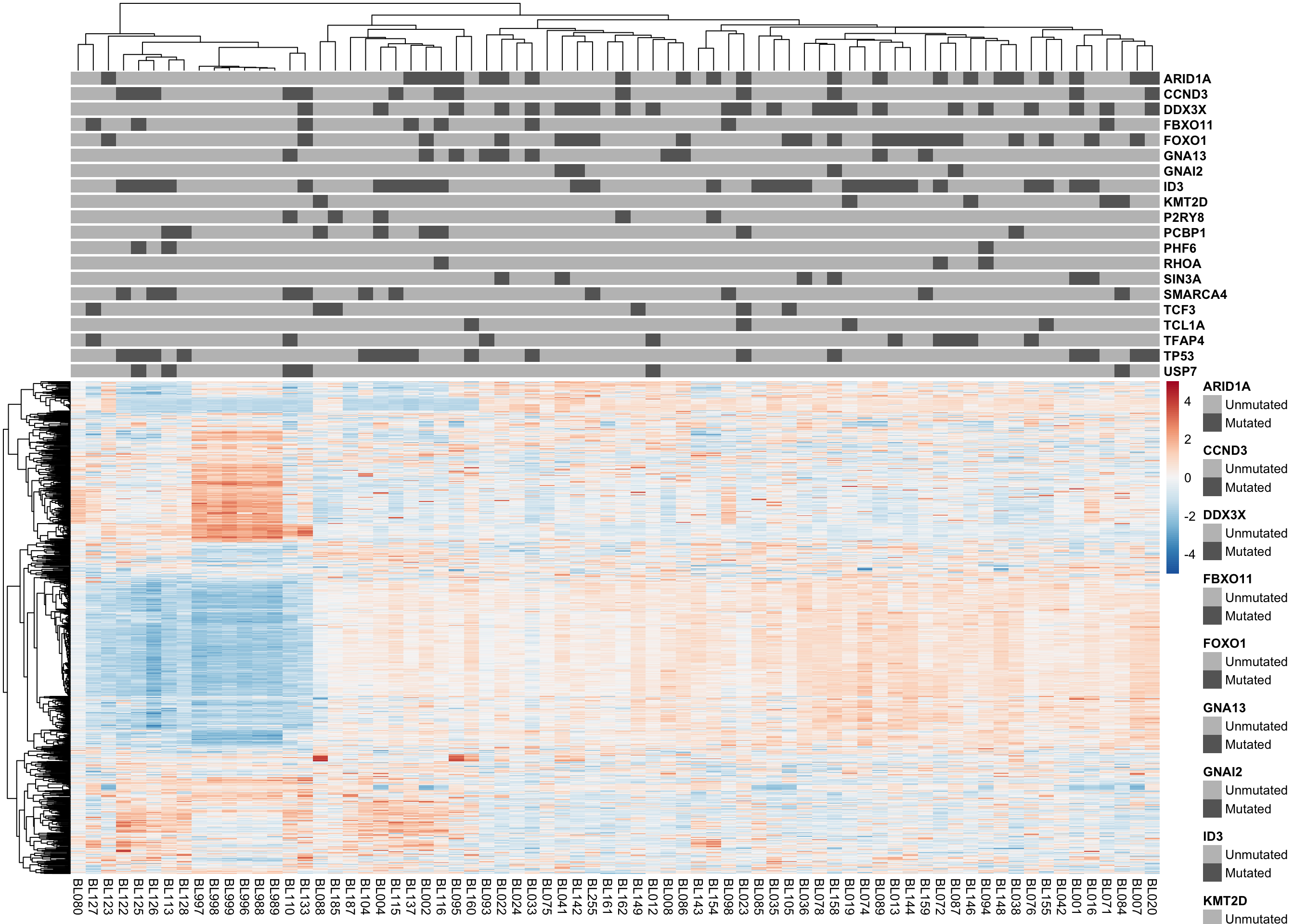
patients <- colnames(salmon$clean$counts)
mut_status <-
maf@data %>%
filter(
grepl("p[.][^=]", HGVSp_Short),
Consequence != "synonymous_variant",
Hugo_Symbol %in% smgs,
patient %in% patients) %>%
select(patient, Hugo_Symbol) %>%
distinct() %>%
mutate(Status = "Mutated") %>%
bind_rows(tibble(patient = patients, Hugo_Symbol = NA)) %>%
spread(Hugo_Symbol, Status, fill = "Unmutated") %>%
select(-`<NA>`) %>%
as.data.frame() %>%
remove_rownames() %>%
column_to_rownames("patient")
mut_colours <- list()
for (gene in names(mut_status)) mut_colours[[gene]] <- c(Unmutated = "grey75", Mutated = "grey40")
plot_heatmap(assay(most_var(salmon$clean$cvst, ntop)), mut_colours, metadata = mut_status)
maf_gr <-
maf@data %>%
makeGRangesFromDataFrame(keep.extra.columns = TRUE, seqnames.field = "Chromosome",
start.field = "Start_Position", end.field = "End_Position")
hits <- findOverlaps(maf_gr, GRanges("chr9", IRanges(37371000, 37372000)))
affected_patients <- maf_gr[queryHits(hits)] %$% unique(patient)
patients <- unique(maf@data$patient)
patient_order <- c(affected_patients, patients[!patients %in% affected_patients])
plot_region_expr("9", "35645518", "39092517", biomart, salmon,
patient_order, gaps_row = length(affected_patients), scale = "column")
maf@data %>%
filter(is_nonsynonymous(Consequence)) %>%
select(patient, Hugo_Symbol) %>%
distinct() %>%
mutate(status = "mutated") %>%
spread(Hugo_Symbol, status, fill = "unmutated") %>%
arrange(match(maf_patients, rownames(colData(salmon$muts$dds))))
