Distributions and lists of all significant differential expression values of the type: SilentIntergenicRegion for comparisons: Amygdala_mRNA_vs_Frontal_mRNA, Amygdala_Ribo_vs_Frontal_Ribo, Amygdala_Ampl_vs_Frontal_Ampl (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Differential expression plots for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: SilentIntergenicRegion
Distribution of log2 differential expression values for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: SilentIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: SilentIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
Scatter plot of log2 expression values for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: SilentIntergenicRegion
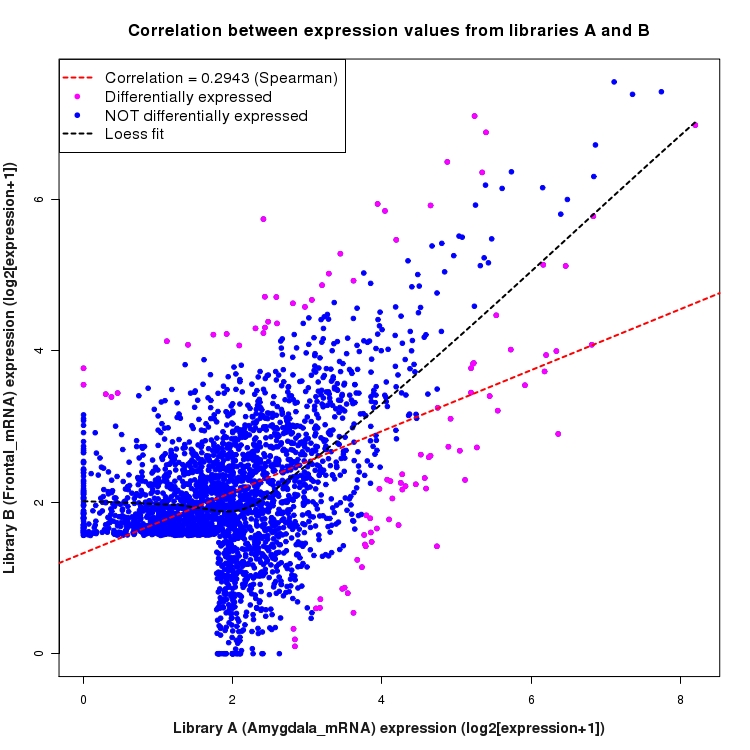
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
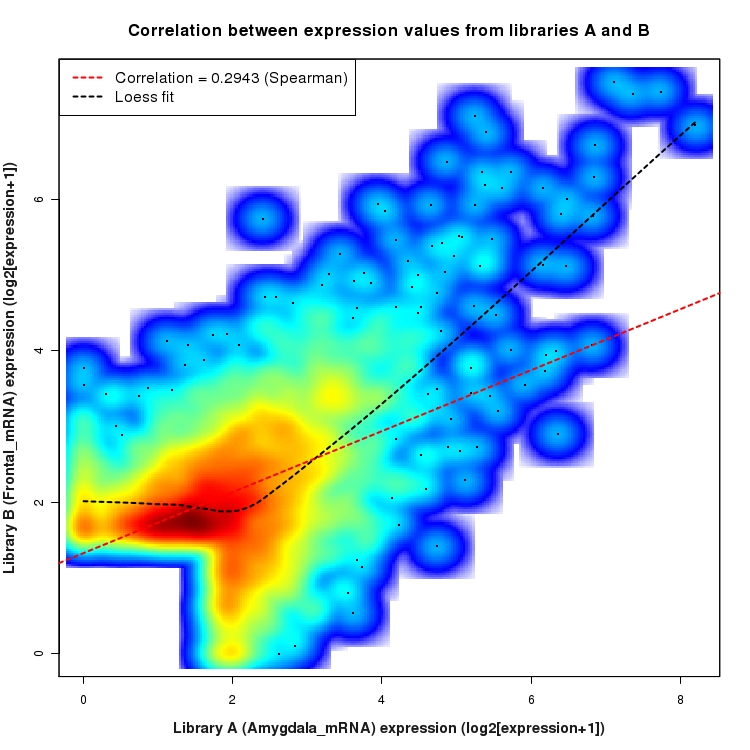
SmoothScatter plot of log2 expression values for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Differential expression plots for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: SilentIntergenicRegion
Distribution of log2 differential expression values for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: SilentIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: SilentIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
Scatter plot of log2 expression values for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: SilentIntergenicRegion
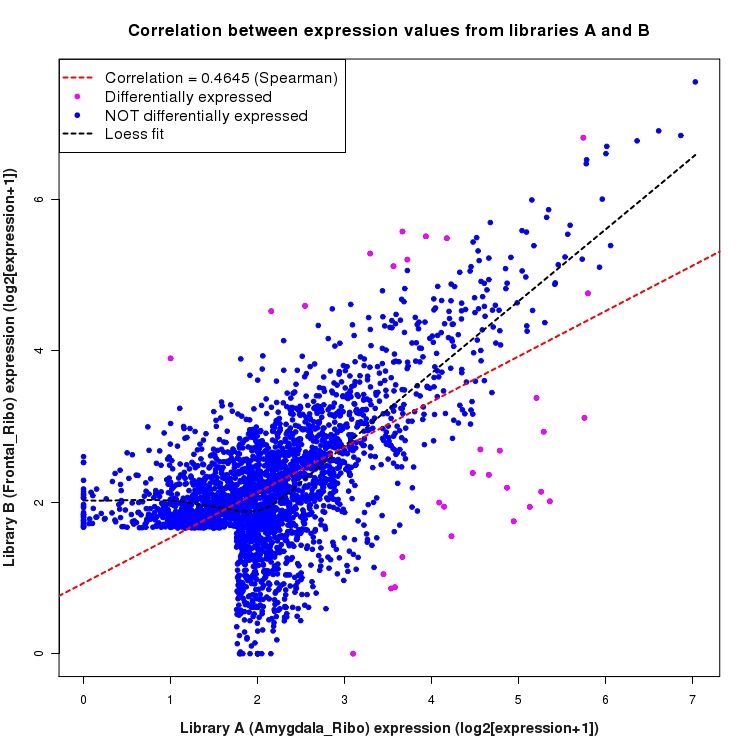
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
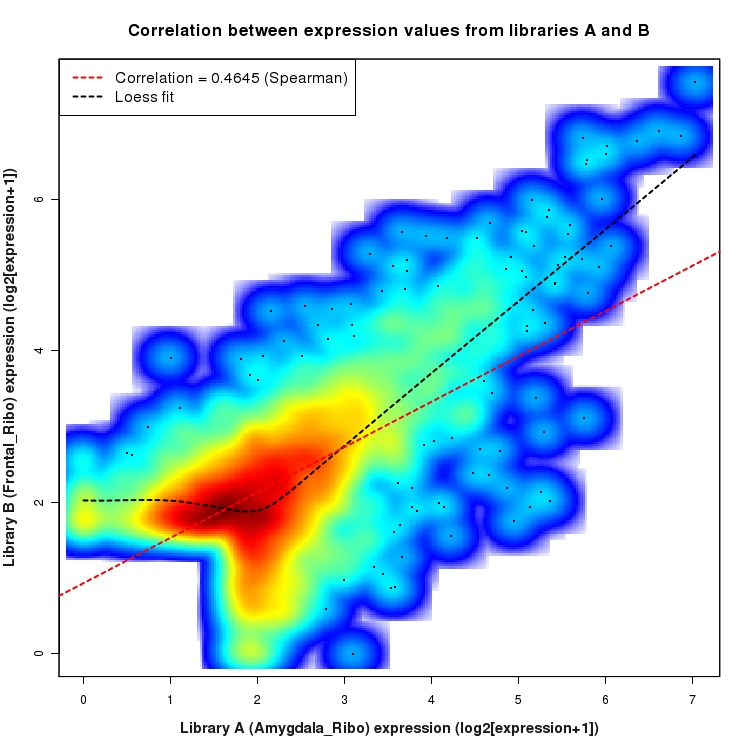
SmoothScatter plot of log2 expression values for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Differential expression plots for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: SilentIntergenicRegion
Distribution of log2 differential expression values for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: SilentIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: SilentIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
Scatter plot of log2 expression values for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
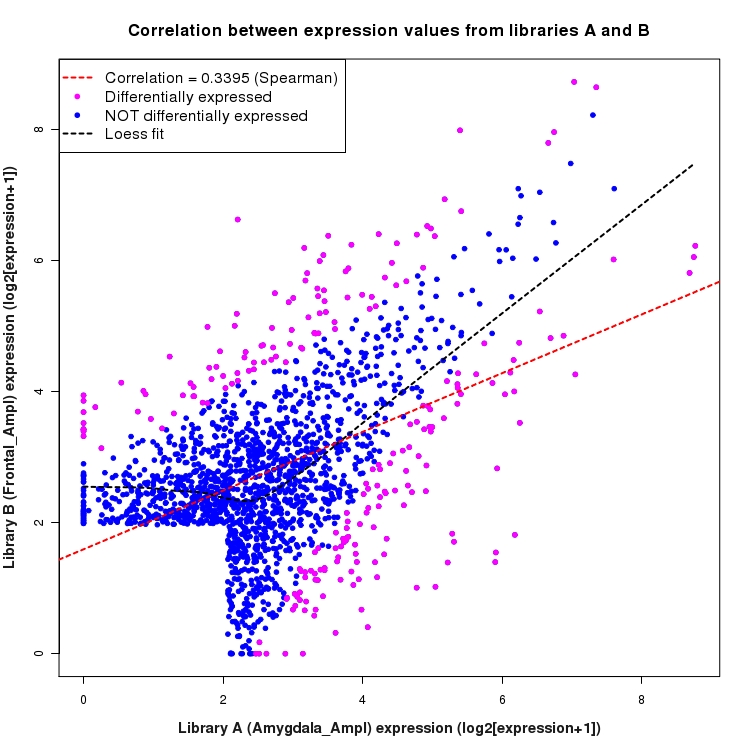
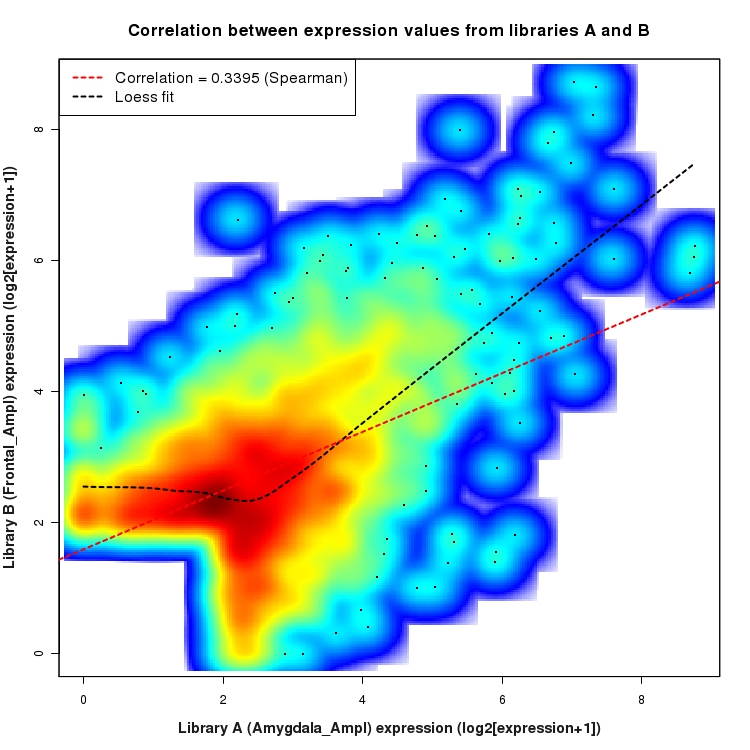
SmoothScatter plot of log2 expression values for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed SilentIntergenicRegion features for pairwise comparisons
The following table provides a ranked list of all significant differentially expressed SilentIntergenicRegion features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | Amygdala_mRNA_vs_Frontal_mRNA (Gene | Feature | Links | Values) | Amygdala_Ribo_vs_Frontal_Ribo (Gene | Feature | Links | Values) | Amygdala_Ampl_vs_Frontal_Ampl (Gene | Feature | Links | Values) |
| 1 | AL390816.1 KCNH5 | IG27_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -13.64 | q.value = 0.00364 | RP11-222A11.1 CTNNA3 | IG36_SR11 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 10.15 | q.value = 1.01e-10 | CLSTN2 TRIM42 | IG39_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 22.61 | q.value = 2.41e-21 |
| 2 | RP11-115M14.1 RP11-316C12.1 | IG42_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -11.73 | q.value = 0.0114 | RP11-222A11.1 CTNNA3 | IG36_SR16 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 9.14 | q.value = 2.65e-08 | MUCL1 KIAA0748 | IG11_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -21.43 | q.value = 1.5e-23 |
| 3 | HSPBP1 BRSK1 | IG14_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 11.02 | q.value = 5.83e-23 | RP11-222A11.1 CTNNA3 | IG36_SR14 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 9.11 | q.value = 3.41e-09 | XRCC5 AC012513.4 | IG13_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 20.61 | q.value = 2.99e-25 |
| 4 | RNF149 CREG2 | IG46_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -10.04 | q.value = 2.15e-11 | RP11-222A11.1 CTNNA3 | IG36_SR15 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 8.68 | q.value = 1.2e-09 | AC015574.2 AC012409.1 | IG30_SR34 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 20.55 | q.value = 2.41e-21 |
| 5 | CRTAP SUSD5 | IG11_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 9.99 | q.value = 1.45e-07 | GAP43 LSAMP | IG49_SR2 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 8.56 | q.value = 0.0201 | SNORD109A SNORD116 | IG7_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 16.22 | q.value = 5.21e-11 |
| 6 | SLIT2 PACRGL | IG19_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -8.75 | q.value = 0.0172 | UBE2V2 AC104989.1 | IG12_SR1 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -7.46 | q.value = 0.0336 | RP11-115M14.1 RP11-316C12.1 | IG42_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -15.40 | q.value = 0.000869 |
| 7 | CHD7 AC022182.1 | IG40_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 8.45 | q.value = 0.00388 | AC116312.1 SH3TC2 | IG21_SR5 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 6.49 | q.value = 0.0256 | OXNAD1 RFTN1 | IG18_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -14.53 | q.value = 0.00139 |
| 8 | PIGG PDE6B | IG7_SR7 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -8.09 | q.value = 0.0255 | RP11-333E5.1 QDPR | IG7_SR6 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 6.39 | q.value = 0.000889 | HHIP ANAPC10 | IG13_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 14.17 | q.value = 8.85e-12 |
| 9 | GS1-256O22.1 SLITRK4 | IG5_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -8.05 | q.value = 0.00205 | RP11-222A11.1 CTNNA3 | IG36_SR12 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 6.38 | q.value = 7.77e-06 | PRH1 TAS2R13 | IG39_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 13.59 | q.value = 1.29e-08 |
| 10 | AL080276.2 RSG17 | IG10_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -7.91 | q.value = 0.0172 | UGT8 SRP_euk_arch | IG25_SR6 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 6.37 | q.value = 0.0256 | AC090805.1 AC022716.1 | IG42_SR24 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -12.89 | q.value = 0.0034 |
| 11 | MAG CD22 | IG19_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 7.07 | q.value = 1.22e-07 | HHIP ANAPC10 | IG13_SR2 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 6.23 | q.value = 5.49e-11 | NKAIN3 AC120042.2 | IG45_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 12.70 | q.value = 1.94e-05 |
| 12 | MAG CD22 | IG19_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 6.70 | q.value = 0.00388 | QKI RP5-826L7.1 | IG4_SR1 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 5.26 | q.value = 0.047 | U6 GRIN2B | IG28_SR11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 12.11 | q.value = 3.9e-12 |
| 13 | C19orf63 JOSD2 | IG47_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 6.67 | q.value = 0.0474 | HSPBP1 BRSK1 | IG14_SR3 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 5.23 | q.value = 0.0329 | AC090805.1 AC022716.1 | IG42_SR23 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -12.10 | q.value = 0.000205 |
| 14 | RP11-222A11.1 CTNNA3 | IG36_SR11 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 6.66 | q.value = 8.08e-24 | MUCL1 KIAA0748 | IG11_SR2 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -5.16 | q.value = 0.00624 | PDE4D U6 | IG39_SR21 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -12.10 | q.value = 0.0022 |
| 15 | ZBTB16 NNMT | IG24_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -6.40 | q.value = 0.0128 | HHIP ANAPC10 | IG13_SR3 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 5.12 | q.value = 1.46e-06 | AC093590.2 AC073043.1 | IG3_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -11.48 | q.value = 0.00835 |
| 16 | RP11-422P15.1 C10orf90 | IG5_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 6.26 | q.value = 0.0474 | QKI RP5-826L7.1 | IG4_SR2 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 4.91 | q.value = 0.000436 | CDC73 RP11-452J13.1 | IG25_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 10.93 | q.value = 8.85e-12 |
| 17 | AC008060.1 AC008060.2 | IG22_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 6.18 | q.value = 0.024 | HHIP ANAPC10 | IG13_SR4 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 4.60 | q.value = 0.00479 | RAP2A RP11-120E13.1 | IG8_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -10.77 | q.value = 0.0129 |
| 18 | ZSCAN2 SCAND2 | IG41_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 6.12 | q.value = 0.024 | AC010130.3 ZEB2 | IG27_SR2 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 4.29 | q.value = 0.000592 | RP11-197K6.1 Y_RNA | IG43_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -10.74 | q.value = 0.0129 |
| 19 | RPL13 CPNE7 | IG43_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 6.02 | q.value = 0.00576 | CNDP1 AC116904.1 | IG29_SR1 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 4.26 | q.value = 0.0334 | GREM1 FMN1 | IG10_SR6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -10.68 | q.value = 0.0129 |
| 20 | U6 GRIN2B | IG28_SR14 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.92 | q.value = 0.0372 | RP11-222A11.1 CTNNA3 | IG36_SR13 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 4.24 | q.value = 0.00441 | AC011288.1 AC005019.2 | IG23_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -10.52 | q.value = 0.0129 |
| 21 | AC116312.1 SH3TC2 | IG21_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.86 | q.value = 1.22e-07 | SERPINI1 RP11-298O21.5 | IG50_SR1 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -4.14 | q.value = 0.0338 | OPCML AP000843.1 | IG7_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -10.00 | q.value = 0.0192 |
| 22 | AC116312.1 SH3TC2 | IG21_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.76 | q.value = 0.000633 | PART1 DEPDC1B | IG43_SR5 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -3.97 | q.value = 0.000985 | MAG CD22 | IG19_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 9.91 | q.value = 4.69e-05 |
| 23 | AC010130.3 ZEB2 | IG27_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.73 | q.value = 0.0372 | GPRIN3 SNCA | IG45_SR18 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -3.76 | q.value = 0.000357 | AC093917.1 PPARGC1A | IG25_SR32 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -9.87 | q.value = 8.02e-05 |
| 24 | U2 | IG1_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.61 | q.value = 0.0474 | HHIP ANAPC10 | IG13_SR1 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 3.63 | q.value = 0.00398 | MKRN3 MAGEL2 | IG66_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 9.80 | q.value = 0.000234 |
| 25 | MUCL1 KIAA0748 | IG11_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -5.55 | q.value = 0.00435 | hsa-mir-138-1 C3orf77 | IG14_SR5 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 3.54 | q.value = 0.000239 | AL390816.1 KCNH5 | IG27_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -9.28 | q.value = 2.28e-06 |
| 26 | MAP2K6 AC003051.1 | IG12_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.49 | q.value = 0.0372 | RP11-307C21.1 FGF12 | IG50_SR5 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -2.99 | q.value = 0.00398 | RP11-160A10.2 RP11-160A10.1 | IG43_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -8.91 | q.value = 0.00412 |
| 27 | KCNJ9 IGSF8 | IG37_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.48 | q.value = 2.28e-13 | GRAMD1B SCN3B | IG2_SR3 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -2.94 | q.value = 0.0335 | ZNF536 AC011478.1 | IG7_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 8.83 | q.value = 0.0035 |
| 28 | CECR2 SLC25A18 | IG8_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.39 | q.value = 0.0152 | NTRK2 RP11-323N23.1 | IG26_SR1 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -2.80 | q.value = 0.0334 | AC016831.2 AC058791.2 | IG22_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 8.55 | q.value = 3.67e-16 |
| 29 | TMEM105 AC110285.3 | IG29_SR13 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.32 | q.value = 0.000265 | GPRIN3 SNCA | IG45_SR19 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -2.48 | q.value = 0.0363 | RP11-115M14.1 RP11-316C12.1 | IG42_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -8.41 | q.value = 0.00412 |
| 30 | PPOX ADAMTS4 | IG18_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.23 | q.value = 0.00395 | SNORD108 AC124312.1 | IG5_SR18 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -2.10 | q.value = 0.000382 | KCNRG DLEU2 | IG28_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 8.21 | q.value = 4.21e-05 |
| 31 | RP11-222A11.1 CTNNA3 | IG36_SR13 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.18 | q.value = 2.99e-11 | PURA C5orf53 | IG24_SR2 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 2.05 | q.value = 0.012 | HECW1 STK17A | IG30_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -8.19 | q.value = 6.91e-13 |
| 32 | TRAPPC9 CHRAC1 | IG2_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.15 | q.value = 6.63e-06 | EMPTY | GABRG1 GABRA2 | IG25_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -7.97 | q.value = 3.56e-06 |
| 33 | RP11-222A11.1 CTNNA3 | IG36_SR14 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.15 | q.value = 0.00756 | EMPTY | MEF2C CETN3 | IG2_SR18 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -7.56 | q.value = 0.0198 |
| 34 | RP11-333E5.1 QDPR | IG7_SR6 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.08 | q.value = 3.54e-08 | EMPTY | AC092111.1 FAM90A1 | IG25_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 7.42 | q.value = 0.0188 |
| 35 | MTMR11 OTUD7B | IG24_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.08 | q.value = 4.92e-14 | EMPTY | UBE2QL1 NSUN2 | IG44_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -7.37 | q.value = 0.0452 |
| 36 | AL138963.1 TPT1 | IG14_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.03 | q.value = 0.00756 | EMPTY | CLSTN2 TRIM42 | IG39_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -7.34 | q.value = 3.74e-14 |
| 37 | C19orf63 JOSD2 | IG47_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 5.01 | q.value = 0.00565 | EMPTY | RP11-222A11.1 CTNNA3 | IG36_SR11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 7.30 | q.value = 6.55e-99 |
| 38 | DYRK1A KCNJ6 | IG20_SR10 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -4.94 | q.value = 0.0154 | EMPTY | AC006112.1 AC007012.1 | IG38_SR12 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -7.16 | q.value = 8.99e-06 |
| 39 | ARHGAP24 MAPK10 | IG7_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -4.86 | q.value = 0.00116 | EMPTY | MAGEH1 USP51 | IG4_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 7.05 | q.value = 0.000523 |
| 40 | SLC22A17 EFS | IG43_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.86 | q.value = 0.00901 | EMPTY | AC010130.3 ZEB2 | IG27_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 7.00 | q.value = 0.000523 |
| 41 | RP11-222A11.1 CTNNA3 | IG36_SR12 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.78 | q.value = 2.47e-12 | EMPTY | ZNF546 ZNF780B | IG16_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.92 | q.value = 3.21e-05 |
| 42 | MAG CD22 | IG19_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.77 | q.value = 0.000818 | EMPTY | RP11-222A11.1 CTNNA3 | IG36_SR15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.88 | q.value = 2.31e-30 |
| 43 | MAL MRPS5 | IG41_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.76 | q.value = 0.0172 | EMPTY | DGKH RP11-215B13.2 | IG35_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.86 | q.value = 0.000159 |
| 44 | TMEM105 AC110285.3 | IG29_SR12 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.65 | q.value = 0.00169 | EMPTY | AC026403.1 AC093304.1 | IG9_SR31 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.79 | q.value = 3.16e-07 |
| 45 | PACRG RP3-495O10.1 | IG2_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.58 | q.value = 0.0172 | EMPTY | DEPDC6 COL14A1 | IG48_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.62 | q.value = 3.56e-17 |
| 46 | TMEM105 AC110285.3 | IG29_SR18 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.47 | q.value = 0.000104 | EMPTY | DAAM1 GPR135 | IG2_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.61 | q.value = 0.00734 |
| 47 | HECW1 STK17A | IG30_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -4.36 | q.value = 0.00388 | EMPTY | RP11-222A11.1 CTNNA3 | IG36_SR12 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.44 | q.value = 9.64e-97 |
| 48 | PPOX ADAMTS4 | IG18_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.31 | q.value = 0.00756 | EMPTY | RP11-124N3.1 CDH18 | IG9_SR6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.33 | q.value = 0.00073 |
| 49 | MTMR11 OTUD7B | IG24_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.30 | q.value = 0.0046 | EMPTY | MMP21 UROS | IG50_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.28 | q.value = 0.00734 |
| 50 | TMEM105 AC110285.3 | IG29_SR10 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.26 | q.value = 0.0087 | EMPTY | ZBTB16 NNMT | IG24_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.28 | q.value = 2.37e-10 |
| 51 | MAG CD22 | IG19_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.16 | q.value = 0.0474 | EMPTY | SNORD113 SNORD113 | IG42_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.15 | q.value = 0.0401 |
| 52 | SLC44A1 FSD1L | IG18_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.12 | q.value = 1.79e-06 | EMPTY | SLC22A9 HRASLS5 | IG32_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.14 | q.value = 0.03 |
| 53 | TRAPPC9 CHRAC1 | IG2_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.11 | q.value = 0.00211 | EMPTY | ESD HTR2A | IG37_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.11 | q.value = 6.56e-10 |
| 54 | U6 C19orf47 | IG25_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.10 | q.value = 0.00211 | EMPTY | GABRG1 GABRA2 | IG25_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.09 | q.value = 1.69e-08 |
| 55 | AC099552.4 INSIG1 | IG19_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.99 | q.value = 0.00756 | EMPTY | RNF149 CREG2 | IG46_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.05 | q.value = 5.54e-38 |
| 56 | ADAMTSL2 FAM163B | IG19_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.99 | q.value = 2e-07 | EMPTY | KIAA1409 PRIMA1 | IG24_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.05 | q.value = 0.00596 |
| 57 | GNB5 | IG1_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.97 | q.value = 0.024 | EMPTY | PHACTR3 U7 | IG36_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 6.01 | q.value = 0.000105 |
| 58 | SLC25A3 AC013283.2 | IG16_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.95 | q.value = 0.0452 | EMPTY | RP11-384F7.1 | IG1_SR9 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.99 | q.value = 0.00115 |
| 59 | TMEM98 SPACA3 | IG48_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.90 | q.value = 0.0474 | EMPTY | USP25 AP001344.1 | IG13_SR6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.91 | q.value = 0.00182 |
| 60 | ESD HTR2A | IG37_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.75 | q.value = 0.037 | EMPTY | TTC7B RPS6KA5 | IG49_SR7 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.87 | q.value = 0.00329 |
| 61 | LSAMP U6 | IG50_SR18 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.74 | q.value = 0.0128 | EMPTY | ARL2BP PLLP | IG11_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.86 | q.value = 0.00238 |
| 62 | KCNJ9 IGSF8 | IG37_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.71 | q.value = 0.00546 | EMPTY | RP11-222A11.1 CTNNA3 | IG36_SR13 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.80 | q.value = 2.29e-92 |
| 63 | WASF3 GPR12 | IG27_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.67 | q.value = 0.024 | EMPTY | TTLL5 TGFB3 | IG14_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.74 | q.value = 0.0263 |
| 64 | RP11-307C21.1 FGF12 | IG50_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.62 | q.value = 6.5e-15 | EMPTY | U6 GRIN2B | IG28_SR15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.73 | q.value = 1.43e-07 |
| 65 | CLSTN2 TRIM42 | IG39_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.58 | q.value = 0.000687 | EMPTY | CUL4A RP11-391H12.1 | IG13_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.72 | q.value = 0.0401 |
| 66 | PPDPF PTK6 | IG38_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.57 | q.value = 0.0236 | EMPTY | AC034110.3 AC034110.2 | IG38_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.72 | q.value = 0.00157 |
| 67 | OPCML AP000843.1 | IG7_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.54 | q.value = 0.0474 | EMPTY | AC138409.3 | IG1_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.68 | q.value = 0.0151 |
| 68 | STK17B HECW2 | IG33_SR3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.53 | q.value = 0.0141 | EMPTY | AC114940.1 EFNA5 | IG43_SR14 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.64 | q.value = 0.00835 |
| 69 | U6 AC092289.1 | IG38_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.53 | q.value = 0.00169 | EMPTY | KIAA0087 AC004947.1 | IG39_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.57 | q.value = 0.0048 |
| 70 | C5orf49 | IG1_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.50 | q.value = 4.58e-06 | EMPTY | SQLE KIAA0196 | IG37_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.54 | q.value = 0.0401 |
| 71 | LLGL2 AC087749.2 | IG31_SR9 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.46 | q.value = 0.0401 | EMPTY | HNRNPA1P4 AC103816.1 | IG24_SR36 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.38 | q.value = 1.13e-05 |
| 72 | AL138963.2 AL138963.1 | IG13_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.42 | q.value = 0.0498 | EMPTY | WASF3 GPR12 | IG27_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.38 | q.value = 4.76e-06 |
| 73 | GABRG1 GABRA2 | IG25_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.40 | q.value = 0.037 | EMPTY | AC116312.1 SH3TC2 | IG21_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.36 | q.value = 3.41e-06 |
| 74 | CNDP1 AC116904.1 | IG29_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.36 | q.value = 0.000275 | EMPTY | ARHGAP24 MAPK10 | IG7_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.30 | q.value = 1.95e-10 |
| 75 | NTRK2 RP11-323N23.1 | IG26_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.32 | q.value = 0.00576 | EMPTY | MAK10 GOLM1 | IG35_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.25 | q.value = 0.0151 |
| 76 | TMEM105 AC110285.3 | IG29_SR16 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.28 | q.value = 6.1e-06 | EMPTY | PACRG RP3-495O10.1 | IG2_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.18 | q.value = 0.00157 |
| 77 | ESD HTR2A | IG37_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.18 | q.value = 0.0105 | EMPTY | BICD1 Y_RNA | IG2_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.18 | q.value = 0.0198 |
| 78 | U6 GRIN2B | IG28_SR8 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.07 | q.value = 3.55e-08 | EMPTY | RP11-451L9.2 RP11-324B6.1 | IG21_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.13 | q.value = 0.0145 |
| 79 | GABRG1 GABRA2 | IG25_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.05 | q.value = 0.0474 | EMPTY | AL359709.1 PREP | IG15_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.12 | q.value = 0.0263 |
| 80 | CCDC8 PNMAL1 | IG2_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.05 | q.value = 0.0265 | EMPTY | GALNT11 RP5-981O7.1 | IG44_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.09 | q.value = 0.0401 |
| 81 | 7SK ZNF706 | IG48_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.83 | q.value = 0.0115 | EMPTY | AP000688.3 RPL23AP3 | IG3_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.09 | q.value = 0.0263 |
| 82 | RP11-307C21.1 FGF12 | IG50_SR4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.81 | q.value = 1.98e-09 | EMPTY | FAM38B AP005117.1 | IG14_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.02 | q.value = 0.03 |
| 83 | AC007283.2 CASP10 | IG17_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.69 | q.value = 0.00364 | EMPTY | RP11-222A11.1 CTNNA3 | IG36_SR16 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 5.00 | q.value = 4.21e-05 |
| 84 | C17orf84 PLD6 | IG43_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.63 | q.value = 0.00482 | EMPTY | GABRG3 AC090696.2 | IG40_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.97 | q.value = 0.0434 |
| 85 | Y_RNA KIF13A | IG45_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.63 | q.value = 0.00576 | EMPTY | PURA C5orf53 | IG24_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.93 | q.value = 0.03 |
| 86 | AGAP1 Y_RNA | IG18_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.54 | q.value = 1.26e-06 | EMPTY | SLC22A25 SLC22A10 | IG30_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.87 | q.value = 0.00262 |
| 87 | FBXO7 LL22NC03-28H9.1 | IG25_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.47 | q.value = 0.0498 | EMPTY | SRP_euk_arch ZNF704 | IG7_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.86 | q.value = 0.0151 |
| 88 | KIAA0664 hsa-mir-1253 | IG49_SR5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.42 | q.value = 0.00881 | EMPTY | LMO4 RP4-544H6.1 | IG45_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.84 | q.value = 0.000761 |
| 89 | GPRIN3 SNCA | IG45_SR18 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.41 | q.value = 0.00121 | EMPTY | NPY5R AC115113.1 | IG28_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.73 | q.value = 0.0102 |
| 90 | NFIC C19orf77 | IG36_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.33 | q.value = 4.2e-20 | EMPTY | CXorf51 RP1-73A14.1 | IG33_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.72 | q.value = 7.99e-05 |
| 91 | NFIC C19orf77 | IG36_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.09 | q.value = 0.0216 | EMPTY | LAYN SIK2 | IG37_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.70 | q.value = 0.0196 |
| 92 | AGAP1 Y_RNA | IG18_SR2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.08 | q.value = 6.1e-06 | EMPTY | GS1-256O22.1 SLITRK4 | IG5_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.68 | q.value = 4.4e-06 |
| 93 | C9orf172 PHPT1 | IG37_SR1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.04 | q.value = 0.00211 | EMPTY | EVI5 RPL5 | IG50_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.68 | q.value = 0.00125 |
| 94 | DYRK1A KCNJ6 | IG20_SR9 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.01 | q.value = 0.00277 | EMPTY | MUDENG NAA30 | IG32_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.68 | q.value = 0.00407 |
| 95 | EMPTY | EMPTY | EED AP003084.2 | IG23_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.67 | q.value = 0.0129 |
| 96 | EMPTY | EMPTY | FBXO7 LL22NC03-28H9.1 | IG25_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.65 | q.value = 0.00575 |
| 97 | EMPTY | EMPTY | UBTF AC003102.2 | IG48_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.58 | q.value = 0.03 |
| 98 | EMPTY | EMPTY | MYL12B RP11-838N2.2 | IG24_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.57 | q.value = 0.0196 |
| 99 | EMPTY | EMPTY | U70984.1 FANCB | IG23_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.53 | q.value = 0.0102 |
| 100 | EMPTY | EMPTY | ADRB1 AC022023.1 | IG22_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.53 | q.value = 5.35e-10 |
| 101 | EMPTY | EMPTY | RP1-313I6.2 RP1-265C24.1 | IG45_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.52 | q.value = 0.00667 |
| 102 | EMPTY | EMPTY | KCNJ2 AC005181.1 | IG17_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.49 | q.value = 1.04e-12 |
| 103 | EMPTY | EMPTY | 7SK ZNF706 | IG48_SR11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.48 | q.value = 0.0385 |
| 104 | EMPTY | EMPTY | USP25 AP001344.1 | IG13_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.45 | q.value = 0.0029 |
| 105 | EMPTY | EMPTY | AC005829.2 LRRC37A2 | IG44_SR42 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.42 | q.value = 0.0102 |
| 106 | EMPTY | EMPTY | TCTEX1D1 | IG1_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.41 | q.value = 0.0029 |
| 107 | EMPTY | EMPTY | AC026357.1 SYT10 | IG8_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.37 | q.value = 0.0362 |
| 108 | EMPTY | EMPTY | FUT9 SRP_euk_arch | IG13_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.32 | q.value = 0.0102 |
| 109 | EMPTY | EMPTY | AC110088.1 AC006427.1 | IG3_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.32 | q.value = 0.03 |
| 110 | EMPTY | EMPTY | RNF32 LMBR1 | IG35_SR8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.30 | q.value = 2.34e-05 |
| 111 | EMPTY | EMPTY | SRP_euk_arch CBLN2 | IG17_SR33 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.28 | q.value = 3.21e-05 |
| 112 | EMPTY | EMPTY | RP11-271F18.1 RP1-186E20.1 | IG22_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.26 | q.value = 1.37e-06 |
| 113 | EMPTY | EMPTY | NCRNA00086 5S_rRNA | IG10_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.26 | q.value = 0.0196 |
| 114 | EMPTY | EMPTY | GS1-256O22.1 SLITRK4 | IG5_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.24 | q.value = 8.5e-07 |
| 115 | EMPTY | EMPTY | ADRB1 AC022023.1 | IG22_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.23 | q.value = 0.0205 |
| 116 | EMPTY | EMPTY | ATP2B1 AC009522.1 | IG38_SR11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.23 | q.value = 0.000147 |
| 117 | EMPTY | EMPTY | AC012317.1 TNRC6A | IG11_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.22 | q.value = 7.56e-11 |
| 118 | EMPTY | EMPTY | RP3-402H5.1 RP11-168F24.1 | IG41_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.21 | q.value = 0.03 |
| 119 | EMPTY | EMPTY | C12orf24 VPS29 | IG38_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.17 | q.value = 0.03 |
| 120 | EMPTY | EMPTY | AC105053.3 POLR1A | IG11_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.15 | q.value = 0.0208 |
| 121 | EMPTY | EMPTY | SCN8A AC068987.1 | IG21_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.14 | q.value = 0.0362 |
| 122 | EMPTY | EMPTY | MAG CD22 | IG19_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.11 | q.value = 6.47e-05 |
| 123 | EMPTY | EMPTY | RP11-333E5.1 QDPR | IG7_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.09 | q.value = 0.00769 |
| 124 | EMPTY | EMPTY | AC087521.5 AC087521.3 | IG40_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.08 | q.value = 0.0208 |
| 125 | EMPTY | EMPTY | CNTN3 RP11-705F7.1 | IG37_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.06 | q.value = 0.0401 |
| 126 | EMPTY | EMPTY | U6 AC092289.1 | IG38_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.06 | q.value = 6.6e-19 |
| 127 | EMPTY | EMPTY | AC015712.1 ALDH1A3 | IG9_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.03 | q.value = 0.0385 |
| 128 | EMPTY | EMPTY | hsa-mir-568 ZBTB20 | IG46_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.02 | q.value = 0.00769 |
| 129 | EMPTY | EMPTY | AL365361.1 AL365361.2 | IG7_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.01 | q.value = 0.00769 |
| 130 | EMPTY | EMPTY | TTC9C ZBTB3 | IG7_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.01 | q.value = 0.00769 |
| 131 | EMPTY | EMPTY | ORC3L AKIRIN2 | IG2_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 4.00 | q.value = 0.0129 |
| 132 | EMPTY | EMPTY | C11orf44 SNX19 | IG49_SR20 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.99 | q.value = 0.0196 |
| 133 | EMPTY | EMPTY | MYL12B RP11-838N2.2 | IG24_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.98 | q.value = 0.0196 |
| 134 | EMPTY | EMPTY | LMAN1 CCBE1 | IG11_SR8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.95 | q.value = 0.00667 |
| 135 | EMPTY | EMPTY | UBE2J1 RRAGD | IG26_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.94 | q.value = 0.00183 |
| 136 | EMPTY | EMPTY | ARL2BP PLLP | IG11_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.92 | q.value = 0.00101 |
| 137 | EMPTY | EMPTY | MAG CD22 | IG19_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.90 | q.value = 0.0208 |
| 138 | EMPTY | EMPTY | AC097634.2 EIF4E3 | IG9_SR11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.89 | q.value = 0.00223 |
| 139 | EMPTY | EMPTY | GABRG1 GABRA2 | IG25_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.89 | q.value = 0.0121 |
| 140 | EMPTY | EMPTY | UBE2V2 AC104989.1 | IG12_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.88 | q.value = 0.0401 |
| 141 | EMPTY | EMPTY | C9orf5 C9orf4 | IG14_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.87 | q.value = 0.0154 |
| 142 | EMPTY | EMPTY | AC026403.1 AC093304.1 | IG9_SR38 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.81 | q.value = 0.000205 |
| 143 | EMPTY | EMPTY | AC113137.1 U6 | IG27_SR10 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.81 | q.value = 0.0385 |
| 144 | EMPTY | EMPTY | RP11-556K13.1 OLFM3 | IG13_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.77 | q.value = 0.000869 |
| 145 | EMPTY | EMPTY | DYRK1A KCNJ6 | IG20_SR9 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.74 | q.value = 0.0154 |
| 146 | EMPTY | EMPTY | MAL MRPS5 | IG41_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.74 | q.value = 0.00596 |
| 147 | EMPTY | EMPTY | SALL1 AC084058.1 | IG13_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.68 | q.value = 0.00476 |
| 148 | EMPTY | EMPTY | RP3-406A7.2 MAP7 | IG14_SR7 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.66 | q.value = 1.16e-14 |
| 149 | EMPTY | EMPTY | NPY5R AC115113.1 | IG28_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.64 | q.value = 0.021 |
| 150 | EMPTY | EMPTY | AL356273.2 IVNS1ABP | IG16_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.62 | q.value = 0.00327 |
| 151 | EMPTY | EMPTY | RP11-545E8.1 A2BP1 | IG42_SR14 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.62 | q.value = 0.00596 |
| 152 | EMPTY | EMPTY | AC093388.1 NAB1 | IG3_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.61 | q.value = 0.0189 |
| 153 | EMPTY | EMPTY | MFI2 DLG1 | IG22_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.61 | q.value = 0.0385 |
| 154 | EMPTY | EMPTY | snoU13 HERC2P3 | IG27_SR6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.58 | q.value = 0.00476 |
| 155 | EMPTY | EMPTY | ANKRD12 | IG1_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.56 | q.value = 0.000738 |
| 156 | EMPTY | EMPTY | QKI RP5-826L7.1 | IG4_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.54 | q.value = 1.06e-09 |
| 157 | EMPTY | EMPTY | U6 GRIN2B | IG28_SR8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.52 | q.value = 0.00122 |
| 158 | EMPTY | EMPTY | ZNF540 ZFP30 | IG43_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.49 | q.value = 0.0385 |
| 159 | EMPTY | EMPTY | SLC22A9 HRASLS5 | IG32_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.48 | q.value = 0.0158 |
| 160 | EMPTY | EMPTY | RP4-774E4.1 KCNA4 | IG19_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.44 | q.value = 0.000568 |
| 161 | EMPTY | EMPTY | OPA1 AC069421.1 | IG6_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.44 | q.value = 0.0294 |
| 162 | EMPTY | EMPTY | FBXO7 LL22NC03-28H9.1 | IG25_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.43 | q.value = 5.03e-07 |
| 163 | EMPTY | EMPTY | SNORD107 SNORD64 | IG3_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.42 | q.value = 0.0158 |
| 164 | EMPTY | EMPTY | MTMR11 OTUD7B | IG24_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.40 | q.value = 0.0358 |
| 165 | EMPTY | EMPTY | FBXO7 LL22NC03-28H9.1 | IG25_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.40 | q.value = 7.74e-11 |
| 166 | EMPTY | EMPTY | SERPINC1 | IG1_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.40 | q.value = 0.0189 |
| 167 | EMPTY | EMPTY | GAS2 AC006299.1 | IG44_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.40 | q.value = 0.0428 |
| 168 | EMPTY | EMPTY | CEP78 PSAT1 | IG14_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.37 | q.value = 0.0428 |
| 169 | EMPTY | EMPTY | OPN5 RP3-402H5.1 | IG40_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.35 | q.value = 0.00158 |
| 170 | EMPTY | EMPTY | ORAI1 MORN3 | IG12_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.31 | q.value = 1.49e-07 |
| 171 | EMPTY | EMPTY | RP11-307C21.1 FGF12 | IG50_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.25 | q.value = 1.23e-35 |
| 172 | EMPTY | EMPTY | CNDP1 AC116904.1 | IG29_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.20 | q.value = 3.72e-09 |
| 173 | EMPTY | EMPTY | AL356131.1 KHDRBS2 | IG13_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.15 | q.value = 0.00145 |
| 174 | EMPTY | EMPTY | EVI5 RPL5 | IG50_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.12 | q.value = 0.00769 |
| 175 | EMPTY | EMPTY | RIMS2 TM7SF4 | IG36_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.08 | q.value = 1.16e-06 |
| 176 | EMPTY | EMPTY | 7SK ZNF706 | IG48_SR8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.08 | q.value = 0.0295 |
| 177 | EMPTY | EMPTY | SNORD108 AC124312.1 | IG5_SR15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.05 | q.value = 3.66e-07 |
| 178 | EMPTY | EMPTY | SOX7 XKR6 | IG17_SR10 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.01 | q.value = 0.0145 |
| 179 | EMPTY | EMPTY | HNRNPA1P4 AC103816.1 | IG24_SR38 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.00 | q.value = 0.019 |
| 180 | EMPTY | EMPTY | U6 GPR37 | IG37_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.00 | q.value = 0.00077 |
| 181 | EMPTY | EMPTY | AC073534.1 AC006504.1 | IG42_SR7 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.98 | q.value = 0.0227 |
| 182 | EMPTY | EMPTY | CACNA1E 7SK | IG18_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.98 | q.value = 6.71e-22 |
| 183 | EMPTY | EMPTY | NTS MGAT4C | IG22_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.96 | q.value = 0.000247 |
| 184 | EMPTY | EMPTY | AC008413.1 DRD1 | IG16_SR7 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.96 | q.value = 0.019 |
| 185 | EMPTY | EMPTY | QKI RP5-826L7.1 | IG4_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.95 | q.value = 0.0196 |
| 186 | EMPTY | EMPTY | C1orf146 GLMN | IG45_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.94 | q.value = 0.0461 |
| 187 | EMPTY | EMPTY | SHISA9 ERCC4 | IG46_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.94 | q.value = 0.0385 |
| 188 | EMPTY | EMPTY | KCNJ3 AC093112.1 | IG41_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.93 | q.value = 8.02e-05 |
| 189 | EMPTY | EMPTY | RPS26P6 YWHAZ | IG45_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.91 | q.value = 8.23e-05 |
| 190 | EMPTY | EMPTY | RPL13 CPNE7 | IG43_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.90 | q.value = 0.000978 |
| 191 | EMPTY | EMPTY | SNORD108 AC124312.1 | IG5_SR10 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.88 | q.value = 0.019 |
| 192 | EMPTY | EMPTY | SCRG1 RP11-489G11.1 | IG24_SR6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.87 | q.value = 0.0461 |
| 193 | EMPTY | EMPTY | PART1 DEPDC1B | IG43_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.86 | q.value = 1.7e-06 |
| 194 | EMPTY | EMPTY | 7SK ZNF706 | IG48_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.85 | q.value = 0.00529 |
| 195 | EMPTY | EMPTY | hsa-mir-125b-1 BLID | IG31_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.82 | q.value = 3.92e-08 |
| 196 | EMPTY | EMPTY | RP1-177G6.1 AL078639.1 | IG28_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.81 | q.value = 0.00148 |
| 197 | EMPTY | EMPTY | C5orf49 | IG1_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.81 | q.value = 0.0042 |
| 198 | EMPTY | EMPTY | NSF WNT3 | IG46_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.79 | q.value = 0.012 |
| 199 | EMPTY | EMPTY | SHISA9 ERCC4 | IG46_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.77 | q.value = 0.00786 |
| 200 | EMPTY | EMPTY | U6 RP11-545E8.1 | IG41_SR8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.77 | q.value = 0.0129 |
| 201 | EMPTY | EMPTY | DNAJB5 RP11-392A14.2 | IG5_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.74 | q.value = 0.0029 |
| 202 | EMPTY | EMPTY | SNORD116 SNORD115 | IG27_SR15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.74 | q.value = 0.0394 |
| 203 | EMPTY | EMPTY | KAT2B SGOL1 | IG32_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.72 | q.value = 0.000278 |
| 204 | EMPTY | EMPTY | U8 FBXL7 | IG36_SR44 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.70 | q.value = 0.0206 |
| 205 | EMPTY | EMPTY | AC103816.1 U6 | IG25_SR8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.69 | q.value = 0.0013 |
| 206 | EMPTY | EMPTY | DEPDC6 COL14A1 | IG48_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.60 | q.value = 0.0151 |
| 207 | EMPTY | EMPTY | CNTNAP4 MON1B | IG28_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.56 | q.value = 0.000114 |
| 208 | EMPTY | EMPTY | OC90 KCNQ3 | IG23_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.56 | q.value = 0.0278 |
| 209 | EMPTY | EMPTY | DPYSL3 JAKMIP2 | IG9_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.54 | q.value = 4.63e-06 |
| 210 | EMPTY | EMPTY | AC009707.1 SNORA70 | IG18_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.54 | q.value = 9.35e-05 |
| 211 | EMPTY | EMPTY | AL451131.1 AGTPBP1 | IG29_SR7 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.54 | q.value = 0.0282 |
| 212 | EMPTY | EMPTY | MAG CD22 | IG19_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.51 | q.value = 0.0319 |
| 213 | EMPTY | EMPTY | SHISA9 ERCC4 | IG46_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.50 | q.value = 0.00844 |
| 214 | EMPTY | EMPTY | U6 GRIN2B | IG28_SR9 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.48 | q.value = 1.97e-08 |
| 215 | EMPTY | EMPTY | SNORD108 AC124312.1 | IG5_SR16 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.47 | q.value = 1.17e-21 |
| 216 | EMPTY | EMPTY | AC127070.1 ZNF605 | IG19_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.46 | q.value = 0.000577 |
| 217 | EMPTY | EMPTY | MRPS28 TPD52 | IG48_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.36 | q.value = 0.0017 |
| 218 | EMPTY | EMPTY | 7SK ZNF706 | IG48_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.36 | q.value = 0.00769 |
| 219 | EMPTY | EMPTY | RP1-177G6.1 AL078639.1 | IG28_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.35 | q.value = 0.0145 |
| 220 | EMPTY | EMPTY | C5orf33 RANBP3L | IG18_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.34 | q.value = 4.79e-12 |
| 221 | EMPTY | EMPTY | AC126365.1 CCDC144NL | IG9_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.26 | q.value = 0.0173 |
| 222 | EMPTY | EMPTY | NTRK2 RP11-323N23.1 | IG26_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.24 | q.value = 0.0362 |
| 223 | EMPTY | EMPTY | Y_RNA KIF13A | IG45_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.22 | q.value = 0.00769 |
| 224 | EMPTY | EMPTY | KCTD16 AC008716.2 | IG42_SR1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.21 | q.value = 0.0173 |
| 225 | EMPTY | EMPTY | RP11-307C21.1 FGF12 | IG50_SR5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.21 | q.value = 1.78e-09 |
| 226 | EMPTY | EMPTY | RNFT2 HRK | IG46_SR4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.17 | q.value = 0.0401 |
| 227 | EMPTY | EMPTY | E2F8 5S_rRNA | IG31_SR18 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.12 | q.value = 0.00407 |
| 228 | EMPTY | EMPTY | CLSTN2 TRIM42 | IG39_SR2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.11 | q.value = 0.0245 |
| 229 | EMPTY | EMPTY | AC104759.3 AC104759.2 | IG37_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.09 | q.value = 0.0295 |
| 230 | EMPTY | EMPTY | PNMA2 DPYSL2 | IG50_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.06 | q.value = 0.00767 |
| 231 | EMPTY | EMPTY | STK17B HECW2 | IG33_SR3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.04 | q.value = 0.019 |
| 232 | EMPTY | EMPTY | DPYD MIR137 | IG20_SR8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.01 | q.value = 0.0012 |