Distributions and lists of all significant differential expression values of the type: Intergenic for comparisons: Amygdala_mRNA_vs_Frontal_mRNA, Amygdala_Ribo_vs_Frontal_Ribo, Amygdala_Ampl_vs_Frontal_Ampl (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Differential expression plots for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: Intergenic
Distribution of log2 differential expression values for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
Scatter plot of log2 expression values for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: Intergenic
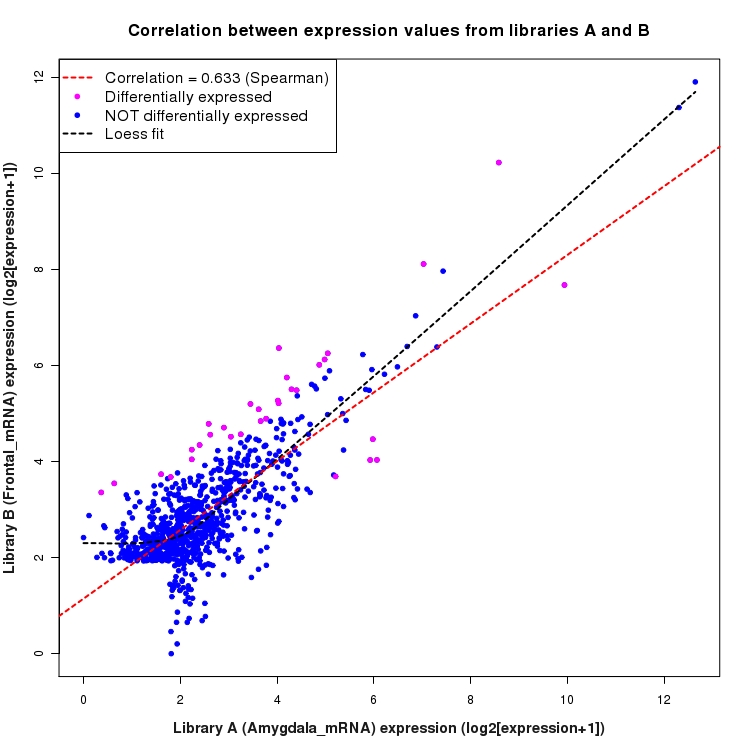
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
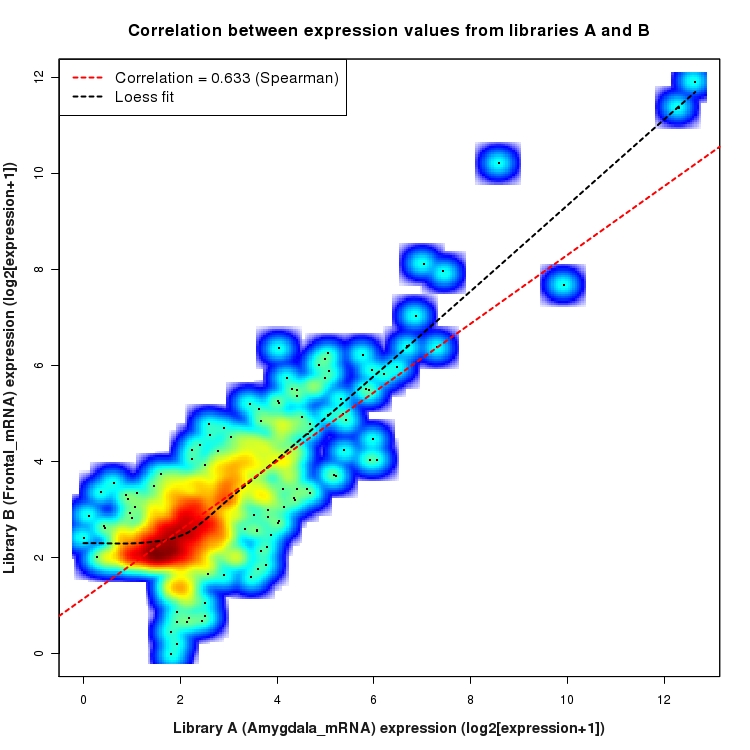
SmoothScatter plot of log2 expression values for comparison: Amygdala_mRNA_vs_Frontal_mRNA and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Differential expression plots for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: Intergenic
Distribution of log2 differential expression values for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
Scatter plot of log2 expression values for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: Intergenic
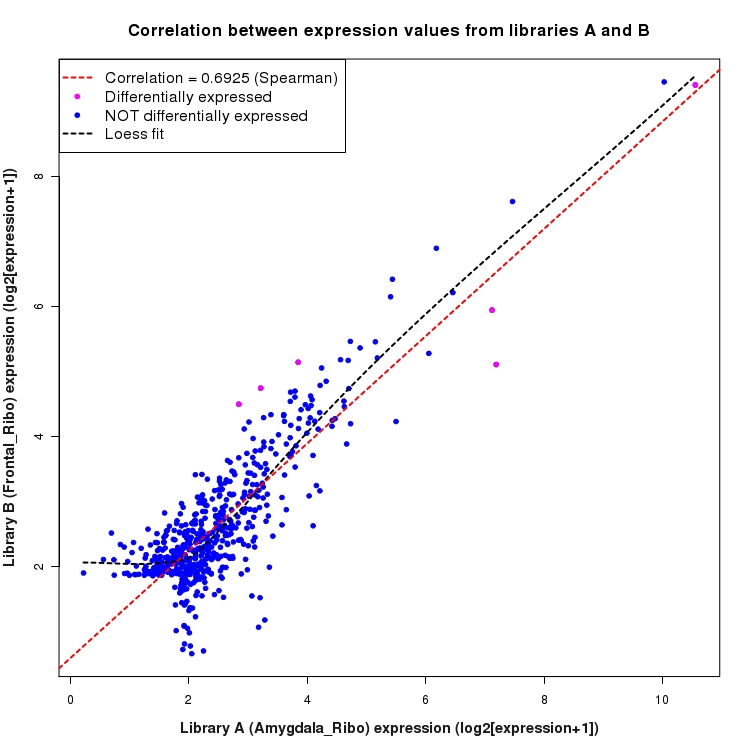
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
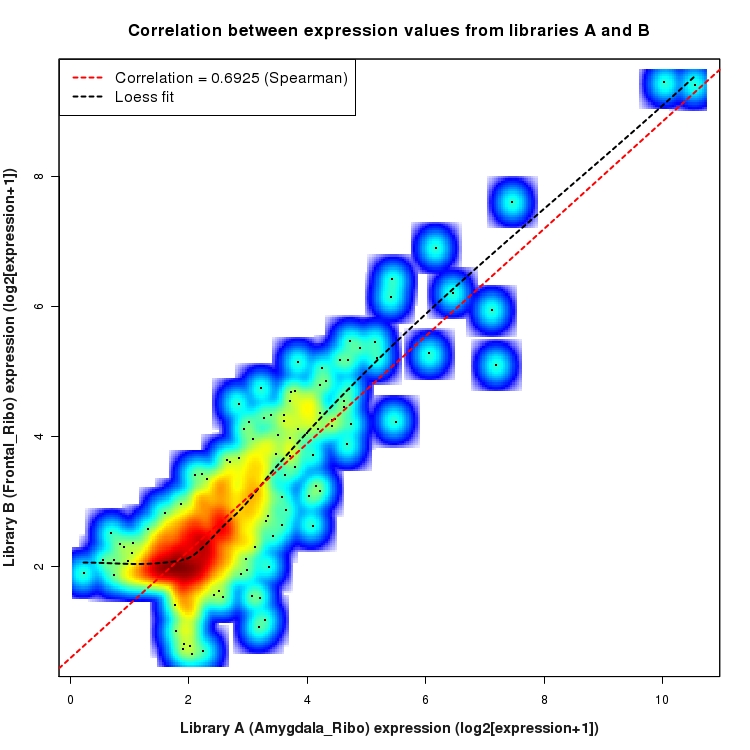
SmoothScatter plot of log2 expression values for comparison: Amygdala_Ribo_vs_Frontal_Ribo and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Differential expression plots for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: Intergenic
Distribution of log2 differential expression values for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
Scatter plot of log2 expression values for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
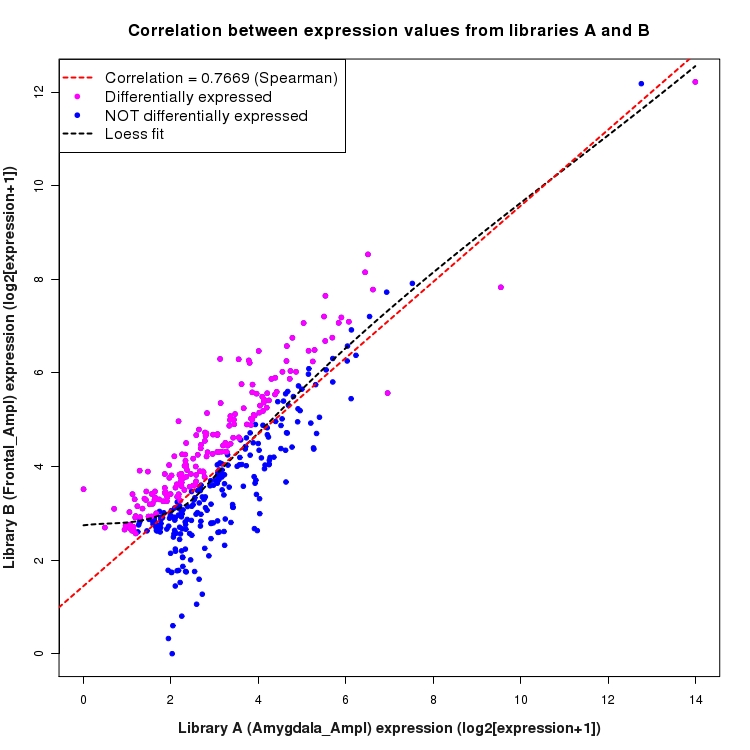
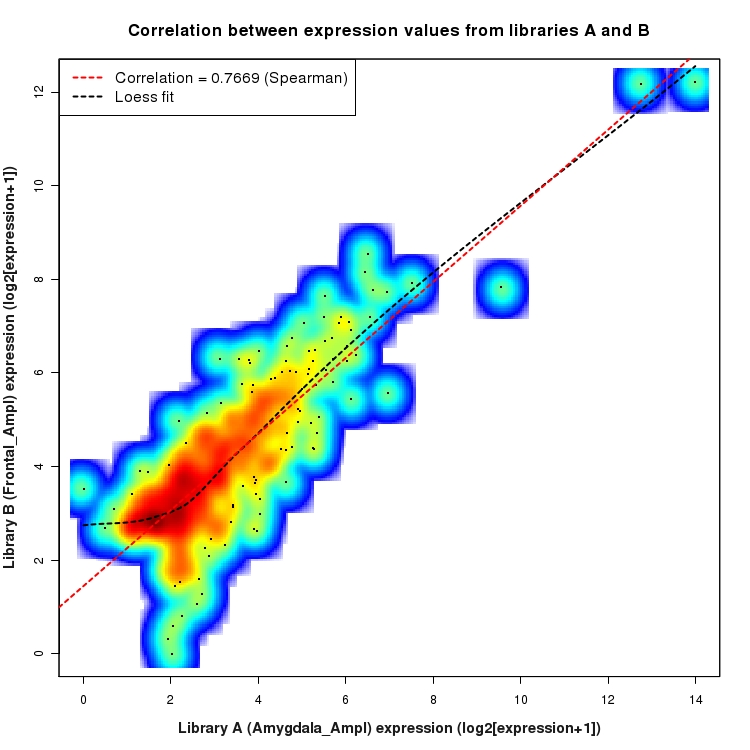
SmoothScatter plot of log2 expression values for comparison: Amygdala_Ampl_vs_Frontal_Ampl and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed Intergenic features for pairwise comparisons
The following table provides a ranked list of all significant differentially expressed Intergenic features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | Amygdala_mRNA_vs_Frontal_mRNA (Gene | Feature | Links | Values) | Amygdala_Ribo_vs_Frontal_Ribo (Gene | Feature | Links | Values) | Amygdala_Ampl_vs_Frontal_Ampl (Gene | Feature | Links | Values) |
| 1 | NCAN HAPLN4 | IG32 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -7.95 | q.value = 0.0165 | MAP4K4 AC005035.1 | IG2 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 4.25 | q.value = 9.3e-17 | TSFM AVIL | IG19 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -11.43 | q.value = 0.00021 |
| 2 | C6orf91 REPS1 | IG43 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -7.52 | q.value = 0.0299 | ACAA2 SCARNA17 | IG48 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -3.14 | q.value = 0.0186 | RP3-525N10.1 BAI3 | IG46 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -9.03 | q.value = 5.63e-23 |
| 3 | C5orf49 | IG1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -5.02 | q.value = 1.94e-14 | C1orf192 RP11-122G18.1 | IG25 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -2.89 | q.value = 0.0186 | HHAT KCNH1 | IG30 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.93 | q.value = 8.93e-09 |
| 4 | MAP4K4 AC005035.1 | IG2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.80 | q.value = 4.5e-113 | ARHGAP24 MAPK10 | IG7 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = -2.45 | q.value = 0.00816 | ARHGAP24 MAPK10 | IG7 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.69 | q.value = 1.37e-20 |
| 5 | ARHGAP24 MAPK10 | IG7 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -4.59 | q.value = 0.000234 | RP11-732M18.1 TULP4 | IG6 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 2.26 | q.value = 1.99e-05 | STXBP5 RP11-361F15.1 | IG47 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -6.18 | q.value = 0.000202 |
| 6 | KALRN UMPS | IG17 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -4.39 | q.value = 0.0382 | J01415.22 | IG33 | (Amygdala_Ribo) | (Frontal_Ribo) | FC = 2.21 | q.value = 1.43e-73 | KIF5A AC022506.1 | IG5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.60 | q.value = 1.92e-18 |
| 7 | MTMR11 OTUD7B | IG24 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 4.08 | q.value = 3.15e-06 | EMPTY | PTPN4 AC016691.1 | IG25 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.50 | q.value = 1.96e-21 |
| 8 | PTPN4 AC016691.1 | IG25 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -4.03 | q.value = 0.0123 | EMPTY | Y_RNA C13orf1 | IG26 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.35 | q.value = 0.000202 |
| 9 | SLC25A3 AC013283.2 | IG16 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.85 | q.value = 0.00537 | EMPTY | C5orf49 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.34 | q.value = 1.96e-17 |
| 10 | NPM2 FGF17 | IG47 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.84 | q.value = 0.00241 | EMPTY | RP4-610C12.3 RP4-610C12.4 | IG15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -5.26 | q.value = 0.00583 |
| 11 | KCNJ9 IGSF8 | IG37 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 3.71 | q.value = 5.41e-05 | EMPTY | C6orf91 REPS1 | IG43 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.98 | q.value = 1.99e-08 |
| 12 | SLC35F3 RP5-827C21.1 | IG25 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.67 | q.value = 0.0382 | EMPTY | GS1-18A18.3 GS1-18A18.2 | IG35 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.89 | q.value = 0.0013 |
| 13 | SCN8A AC068987.1 | IG21 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.50 | q.value = 0.0417 | EMPTY | SNRPN SNORD107 | IG2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.66 | q.value = 2.12e-09 |
| 14 | FBXO7 LL22NC03-28H9.1 | IG25 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.50 | q.value = 0.00177 | EMPTY | hsa-mir-654 hsa-mir-300 | IG20 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.62 | q.value = 0.0157 |
| 15 | RP3-525N10.1 BAI3 | IG46 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.36 | q.value = 0.000133 | EMPTY | WNK1 RAD52 | IG11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.45 | q.value = 1.73e-05 |
| 16 | KIF5A AC022506.1 | IG5 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -3.12 | q.value = 1.87e-152 | EMPTY | TUBGCP4 TP53BP1 | IG26 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.44 | q.value = 1.37e-06 |
| 17 | GRAMD1B SCN3B | IG2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.93 | q.value = 9.78e-06 | EMPTY | FBXO7 LL22NC03-28H9.1 | IG25 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.43 | q.value = 1.14e-11 |
| 18 | RP11-732M18.1 TULP4 | IG6 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.86 | q.value = 0.0417 | EMPTY | CAND1 AC078983.2 | IG21 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.41 | q.value = 0.000108 |
| 19 | URM1 hsa-mir-219-2 | IG21 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = 2.85 | q.value = 0.00209 | EMPTY | SENP5 NCBP2 | IG17 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.38 | q.value = 0.00276 |
| 20 | SNRPN SNORD107 | IG2 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.77 | q.value = 0.0186 | EMPTY | KLHDC10 AC087071.2 | IG6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.33 | q.value = 5.5e-41 |
| 21 | SNORD64 SNORD108 | IG4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.77 | q.value = 0.00176 | EMPTY | SNORD64 SNORD108 | IG4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.26 | q.value = 1.73e-05 |
| 22 | PARVB PARVG | IG20 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.49 | q.value = 0.0446 | EMPTY | NUDT4 UBE2N | IG12 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.21 | q.value = 0.000309 |
| 23 | TUBGCP4 TP53BP1 | IG26 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.38 | q.value = 0.00348 | EMPTY | MCM4 U6 | IG10 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.09 | q.value = 2.26e-26 |
| 24 | PHACTR1 AL008729.1 | IG45 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.31 | q.value = 5.68e-06 | EMPTY | AC051642.2 AC051642.1 | IG29 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.08 | q.value = 0.00418 |
| 25 | CCDC65 ARF3 | IG49 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.31 | q.value = 0.00176 | EMPTY | SSBP3 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.08 | q.value = 3.14e-73 |
| 26 | SNORD107 SNORD64 | IG3 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.27 | q.value = 0.00635 | EMPTY | PHTF1 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -4.01 | q.value = 0.000108 |
| 27 | GABRB1 COMMD8 | IG29 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.27 | q.value = 0.0342 | EMPTY | ICA1L AC010900.2 | IG4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.95 | q.value = 0.00134 |
| 28 | USP14 THOC1 | IG4 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.21 | q.value = 5.33e-05 | EMPTY | PABPC1L2B PABPC1L2A | IG23 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.94 | q.value = 0.0125 |
| 29 | MARCH6 ROPN1L | IG18 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.21 | q.value = 6.72e-05 | EMPTY | PHACTR1 AL008729.1 | IG45 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.93 | q.value = 3.78e-21 |
| 30 | SSSCA1 FAM89B | IG14 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.17 | q.value = 0.0402 | EMPTY | MPP6 DFNA5 | IG19 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.86 | q.value = 1.73e-05 |
| 31 | SSBP3 | IG1 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.12 | q.value = 2.17e-20 | EMPTY | SCN8A AC068987.1 | IG21 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.81 | q.value = 0.000372 |
| 32 | EFR3B POMC | IG23 | (Amygdala_mRNA) | (Frontal_mRNA) | FC = -2.12 | q.value = 0.00376 | EMPTY | GABRB1 COMMD8 | IG29 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.79 | q.value = 4.71e-18 |
| 33 | EMPTY | EMPTY | VPS13A GNA14 | IG6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.78 | q.value = 0.0157 |
| 34 | EMPTY | EMPTY | TRMT112 PRDX5 | IG10 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.78 | q.value = 0.008 |
| 35 | EMPTY | EMPTY | C1orf146 GLMN | IG45 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.73 | q.value = 1.73e-05 |
| 36 | EMPTY | EMPTY | CHST7 SLC9A7 | IG26 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.69 | q.value = 2.28e-10 |
| 37 | EMPTY | EMPTY | NEURL SH3PXD2A | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.69 | q.value = 1.73e-05 |
| 38 | EMPTY | EMPTY | GALNT9 AC148477.2 | IG8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.64 | q.value = 3.31e-05 |
| 39 | EMPTY | EMPTY | RBM4B SPTBN2 | IG9 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.62 | q.value = 0.008 |
| 40 | EMPTY | EMPTY | DGKE U6 | IG19 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.61 | q.value = 0.00418 |
| 41 | EMPTY | EMPTY | KBTBD10 FASTKD1 | IG41 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.56 | q.value = 0.00418 |
| 42 | EMPTY | EMPTY | SLC25A18 ATP6V1E1 | IG9 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.47 | q.value = 0.00134 |
| 43 | EMPTY | EMPTY | BCL2L13 BID | IG11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.47 | q.value = 0.00134 |
| 44 | EMPTY | EMPTY | WNT2B ST7L | IG37 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.46 | q.value = 0.0013 |
| 45 | EMPTY | EMPTY | ASH2L STAR | IG37 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.43 | q.value = 0.000108 |
| 46 | EMPTY | EMPTY | J01415.18 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.42 | q.value = 0 |
| 47 | EMPTY | EMPTY | PRPS2 GS1-324M7.2 | IG48 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.42 | q.value = 0.0252 |
| 48 | EMPTY | EMPTY | RP11-160A10.2 RP11-160A10.1 | IG43 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.41 | q.value = 0.000116 |
| 49 | EMPTY | EMPTY | AC010900.2 WDR12 | IG5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.37 | q.value = 0.0157 |
| 50 | EMPTY | EMPTY | RP11-324H6.1 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.36 | q.value = 0.0125 |
| 51 | EMPTY | EMPTY | OSGEPL1 ORMDL1 | IG49 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.32 | q.value = 0.0125 |
| 52 | EMPTY | EMPTY | RP1-251M9.1 RP1-251M9.2 | IG26 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.31 | q.value = 1.54e-08 |
| 53 | EMPTY | EMPTY | AP001053.1 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.29 | q.value = 6.27e-05 |
| 54 | EMPTY | EMPTY | AL078639.1 CDR1 | IG29 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.29 | q.value = 2.81e-06 |
| 55 | EMPTY | EMPTY | MAP4K4 AC005035.1 | IG2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 3.29 | q.value = 1.62e-28 |
| 56 | EMPTY | EMPTY | RP4-580O19.1 CSMD2 | IG46 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.28 | q.value = 0.0497 |
| 57 | EMPTY | EMPTY | ACSS3 PPFIA2 | IG9 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.27 | q.value = 1.95e-47 |
| 58 | EMPTY | EMPTY | MPHOSPH8 RP11-523H24.2 | IG55 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.27 | q.value = 0.0013 |
| 59 | EMPTY | EMPTY | AL354798.1 TPTE2P1 | IG8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.27 | q.value = 0.000202 |
| 60 | EMPTY | EMPTY | HSPA4L AC093591.1 | IG42 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.26 | q.value = 0.0023 |
| 61 | EMPTY | EMPTY | ADAM22 SRI | IG27 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.26 | q.value = 0.008 |
| 62 | EMPTY | EMPTY | SNORD107 SNORD64 | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.26 | q.value = 5.85e-25 |
| 63 | EMPTY | EMPTY | TTLL5 TGFB3 | IG14 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.23 | q.value = 0.0252 |
| 64 | EMPTY | EMPTY | SPIN1 RP11-317B17.2 | IG21 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.21 | q.value = 0.0052 |
| 65 | EMPTY | EMPTY | CAMSAP1L1 RPL34P1 | IG23 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.20 | q.value = 0.0157 |
| 66 | EMPTY | EMPTY | ARNTL BTBD10 | IG18 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.16 | q.value = 0.008 |
| 67 | EMPTY | EMPTY | TNFAIP1 POLDIP2 | IG14 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.16 | q.value = 0.0007 |
| 68 | EMPTY | EMPTY | CEP72 AC026740.2 | IG11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.15 | q.value = 4.76e-06 |
| 69 | EMPTY | EMPTY | PDE12 ARF4 | IG40 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.14 | q.value = 0.0023 |
| 70 | EMPTY | EMPTY | FAM92A1 RBM12B | IG33 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.13 | q.value = 1e-05 |
| 71 | EMPTY | EMPTY | DIABLO VPS33A | IG25 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.11 | q.value = 0.000714 |
| 72 | EMPTY | EMPTY | RPS28 KANK3 | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.10 | q.value = 0.0042 |
| 73 | EMPTY | EMPTY | AP4S1 HECTD1 | IG14 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.09 | q.value = 6.27e-05 |
| 74 | EMPTY | EMPTY | RP1-241P17.3 RP1-241P17.1 | IG34 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.08 | q.value = 0.0302 |
| 75 | EMPTY | EMPTY | AL117352.1 EGLN1 | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.08 | q.value = 0.0157 |
| 76 | EMPTY | EMPTY | ITIH2 KIN | IG50 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.08 | q.value = 0.0042 |
| 77 | EMPTY | EMPTY | METTL11A | IG51 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.06 | q.value = 0.000108 |
| 78 | EMPTY | EMPTY | PPIG C2orf77 | IG44 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.06 | q.value = 0.0497 |
| 79 | EMPTY | EMPTY | MPP5 ATP6V1D | IG11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.06 | q.value = 2.85e-12 |
| 80 | EMPTY | EMPTY | KPNA5 FAM162B | IG47 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.05 | q.value = 0.0042 |
| 81 | EMPTY | EMPTY | ZNF549 ZNF550 | IG41 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.04 | q.value = 0.0252 |
| 82 | EMPTY | EMPTY | AL137142.1 HSPH1 | IG43 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -3.03 | q.value = 8.02e-08 |
| 83 | EMPTY | EMPTY | ZNF398 RP4-800G7.2 | IG32 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.99 | q.value = 1.3e-06 |
| 84 | EMPTY | EMPTY | C6orf182 C6orf183 | IG4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.99 | q.value = 0.0151 |
| 85 | EMPTY | EMPTY | MRPL42 AC025260.1 | IG15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.99 | q.value = 0.0151 |
| 86 | EMPTY | EMPTY | NCRNA00086 5S_rRNA | IG10 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.98 | q.value = 9.43e-10 |
| 87 | EMPTY | EMPTY | INTS3 SLC27A3 | IG9 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.97 | q.value = 0.0252 |
| 88 | EMPTY | EMPTY | GMPR ATXN1 | IG33 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.96 | q.value = 0.0253 |
| 89 | EMPTY | EMPTY | SLC24A1 DENND4A | IG6 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.95 | q.value = 1e-05 |
| 90 | EMPTY | EMPTY | ELP4 PAX6 | IG27 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.94 | q.value = 0.0042 |
| 91 | EMPTY | EMPTY | ZDHHC2 CNOT7 | IG16 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.90 | q.value = 2.95e-05 |
| 92 | EMPTY | EMPTY | SNRPF CCDC38 | IG42 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.90 | q.value = 0.0042 |
| 93 | EMPTY | EMPTY | LRCH3 IQCG | IG31 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.87 | q.value = 0.0497 |
| 94 | EMPTY | EMPTY | SNORD113 SNORD113 | IG20 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.85 | q.value = 2.38e-09 |
| 95 | EMPTY | EMPTY | ZNF273 RP11-590C23.1 | IG16 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.85 | q.value = 0.0151 |
| 96 | EMPTY | EMPTY | SKP2 C5orf33 | IG17 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.84 | q.value = 0.00761 |
| 97 | EMPTY | EMPTY | LETM2 FGFR1 | IG45 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.80 | q.value = 0.0135 |
| 98 | EMPTY | EMPTY | CLUAP1 NLRC3 | IG2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.78 | q.value = 9.43e-10 |
| 99 | EMPTY | EMPTY | ST6GALNAC5 RP4-706A16.2 | IG22 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.77 | q.value = 0.0497 |
| 100 | EMPTY | EMPTY | ACOT13 C6orf62 | IG48 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.77 | q.value = 0.0042 |
| 101 | EMPTY | EMPTY | ZC3H14 EML5 | IG30 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.77 | q.value = 0.00115 |
| 102 | EMPTY | EMPTY | SOD3 CCDC149 | IG30 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.74 | q.value = 4.57e-05 |
| 103 | EMPTY | EMPTY | RP5-1024G6.1 LRP8 | IG32 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.74 | q.value = 0.00761 |
| 104 | EMPTY | EMPTY | SNORD113 SNORD113 | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.72 | q.value = 0.0135 |
| 105 | EMPTY | EMPTY | NPB PCYT2 | IG5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.72 | q.value = 0.0302 |
| 106 | EMPTY | EMPTY | PKNOX2 FEZ1 | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.71 | q.value = 0.0302 |
| 107 | EMPTY | EMPTY | SLC25A16 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.70 | q.value = 0.0253 |
| 108 | EMPTY | EMPTY | RFC5 WSB2 | IG4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.70 | q.value = 0.00616 |
| 109 | EMPTY | EMPTY | NAALAD2 AP000648.1 | IG8 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.69 | q.value = 0.00751 |
| 110 | EMPTY | EMPTY | SCRN3 GPR155 | IG45 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.68 | q.value = 0.0253 |
| 111 | EMPTY | EMPTY | CSTF2 NOX1 | IG20 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.68 | q.value = 0.0151 |
| 112 | EMPTY | EMPTY | RP1-12G14.1 RP1-12G14.4 | IG18 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.66 | q.value = 0.00761 |
| 113 | EMPTY | EMPTY | RP11-66D17.1 ISG20L2 | IG50 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.65 | q.value = 7.66e-07 |
| 114 | EMPTY | EMPTY | TBPL1 SLC2A12 | IG38 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.64 | q.value = 0.00185 |
| 115 | EMPTY | EMPTY | KCNAB1 SSR3 | IG22 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = 2.61 | q.value = 0.0013 |
| 116 | EMPTY | EMPTY | ATXN7 RP11-245J9.2 | IG22 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.61 | q.value = 0.00751 |
| 117 | EMPTY | EMPTY | HSP90B1 C12orf73 | IG13 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.60 | q.value = 0.0302 |
| 118 | EMPTY | EMPTY | GNA11 AC005262.1 | IG30 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.59 | q.value = 0.0497 |
| 119 | EMPTY | EMPTY | LONP2 SIAH1 | IG39 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.58 | q.value = 9.71e-07 |
| 120 | EMPTY | EMPTY | MRPL45 GPR179 | IG24 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.55 | q.value = 0.0253 |
| 121 | EMPTY | EMPTY | INPP5F C10orf119 | IG40 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.53 | q.value = 0.0135 |
| 122 | EMPTY | EMPTY | SLC10A5 ZFAND1 | IG20 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.53 | q.value = 0.0302 |
| 123 | EMPTY | EMPTY | U7 C12orf57 | IG38 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.51 | q.value = 7.94e-12 |
| 124 | EMPTY | EMPTY | XXyac-R12DG2.3 AL627107.2 | IG16 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.49 | q.value = 0.00297 |
| 125 | EMPTY | EMPTY | FAM118B SRPR | IG16 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.48 | q.value = 5.46e-06 |
| 126 | EMPTY | EMPTY | SKIL RP11-469J4.3 | IG26 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.48 | q.value = 2.98e-09 |
| 127 | EMPTY | EMPTY | RAD9B PPTC7 | IG39 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.44 | q.value = 6.35e-19 |
| 128 | EMPTY | EMPTY | CACNA1D CHDH | IG18 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.40 | q.value = 2.72e-05 |
| 129 | EMPTY | EMPTY | PSMA3 AL132989.2 | IG40 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.40 | q.value = 0.00126 |
| 130 | EMPTY | EMPTY | GUF1 GNPDA2 | IG20 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.39 | q.value = 0.00143 |
| 131 | EMPTY | EMPTY | Y_RNA MTCH2 | IG44 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.38 | q.value = 0.0017 |
| 132 | EMPTY | EMPTY | NDOR1 RNF208 | IG17 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.37 | q.value = 0.016 |
| 133 | EMPTY | EMPTY | VPS13B COX6C | IG29 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.37 | q.value = 1.42e-05 |
| 134 | EMPTY | EMPTY | SUMF2 PHKG1 | IG15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.36 | q.value = 0.0102 |
| 135 | EMPTY | EMPTY | CIRBP C19orf24 | IG36 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.35 | q.value = 1.39e-16 |
| 136 | EMPTY | EMPTY | SNAPIN ILF2 | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.35 | q.value = 0.0235 |
| 137 | EMPTY | EMPTY | ZNF673 ZNF674 | IG23 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.33 | q.value = 0.00391 |
| 138 | EMPTY | EMPTY | TTC18 ANXA7 | IG30 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.32 | q.value = 0.000103 |
| 139 | EMPTY | EMPTY | CSPP1 ARFGEF1 | IG28 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.31 | q.value = 0.0393 |
| 140 | EMPTY | EMPTY | FAM73A C1orf118 | IG29 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.31 | q.value = 4.55e-11 |
| 141 | EMPTY | EMPTY | AC068768.1 SBNO1 | IG42 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.28 | q.value = 0.00066 |
| 142 | EMPTY | EMPTY | RPL21P44 CHIC2 | IG25 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.28 | q.value = 0.0497 |
| 143 | EMPTY | EMPTY | C6orf89 RP1-90K10.1 | IG41 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.28 | q.value = 0.00019 |
| 144 | EMPTY | EMPTY | AC135050.3 VKORC1 | IG49 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.28 | q.value = 0.0393 |
| 145 | EMPTY | EMPTY | CCDC65 ARF3 | IG49 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.26 | q.value = 2.77e-06 |
| 146 | EMPTY | EMPTY | NHEDC2 BDH2 | IG2 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.26 | q.value = 0.000103 |
| 147 | EMPTY | EMPTY | AP006216.3 AP006216.4 | IG41 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.25 | q.value = 1.62e-05 |
| 148 | EMPTY | EMPTY | AL732292.1 TOMM20 | IG41 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.24 | q.value = 0.000234 |
| 149 | EMPTY | EMPTY | AC009501.2 C2orf86 | IG25 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.24 | q.value = 0.0148 |
| 150 | EMPTY | EMPTY | MSI2 U7 | IG28 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.24 | q.value = 3.87e-08 |
| 151 | EMPTY | EMPTY | RP11-165J3.2 FAM120AOS | IG28 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.24 | q.value = 0.00391 |
| 152 | EMPTY | EMPTY | TENC1 SPRYD3 | IG11 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.23 | q.value = 1.95e-25 |
| 153 | EMPTY | EMPTY | C14orf23 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.23 | q.value = 1.16e-05 |
| 154 | EMPTY | EMPTY | SNORD108 AC124312.1 | IG5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.23 | q.value = 1.96e-07 |
| 155 | EMPTY | EMPTY | TBRG1 SIAE | IG38 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.23 | q.value = 4.02e-12 |
| 156 | EMPTY | EMPTY | PDE8B WDR41 | IG23 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.22 | q.value = 0.002 |
| 157 | EMPTY | EMPTY | ZNF410 FAM161B | IG29 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.21 | q.value = 0.000256 |
| 158 | EMPTY | EMPTY | SF1 MAP4K2 | IG26 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.20 | q.value = 0.0317 |
| 159 | EMPTY | EMPTY | TADA2B GRPEL1 | IG43 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.20 | q.value = 0.0148 |
| 160 | EMPTY | EMPTY | GNAI3 RP5-1160K1.3 | IG31 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.19 | q.value = 0.000108 |
| 161 | EMPTY | EMPTY | PSENEN AD000671.3 | IG47 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.18 | q.value = 0.0042 |
| 162 | EMPTY | EMPTY | ZNF749 VN1R1 | IG36 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.16 | q.value = 0.026 |
| 163 | EMPTY | EMPTY | UBXN2A AC066692.2 | IG15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.13 | q.value = 0.0317 |
| 164 | EMPTY | EMPTY | FAR2 AC009318.1 | IG26 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.12 | q.value = 0.000219 |
| 165 | EMPTY | EMPTY | BAG2 RAB23 | IG48 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.10 | q.value = 0.00136 |
| 166 | EMPTY | EMPTY | AVIL CTDSP2 | IG20 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.10 | q.value = 0.008 |
| 167 | EMPTY | EMPTY | AL136982.2 GLUD1 | IG16 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.10 | q.value = 0.000115 |
| 168 | EMPTY | EMPTY | OFD1 GPM6B | IG15 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.09 | q.value = 7.94e-12 |
| 169 | EMPTY | EMPTY | FAM164C NEK9 | IG3 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.08 | q.value = 0.0393 |
| 170 | EMPTY | EMPTY | AL583832.1 RBBP5 | IG46 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.08 | q.value = 0.0419 |
| 171 | EMPTY | EMPTY | C14orf180 TMEM179 | IG43 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.08 | q.value = 0.000486 |
| 172 | EMPTY | EMPTY | SART3 | IG1 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.05 | q.value = 0.00221 |
| 173 | EMPTY | EMPTY | AC114772.4 GFPT1 | IG36 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.05 | q.value = 0.008 |
| 174 | EMPTY | EMPTY | USP14 THOC1 | IG4 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.03 | q.value = 1.75e-14 |
| 175 | EMPTY | EMPTY | RP11-321E8.4 AC016769.1 | IG5 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.01 | q.value = 0.0344 |
| 176 | EMPTY | EMPTY | TMEM126A CREBZF | IG14 | (Amygdala_Ampl) | (Frontal_Ampl) | FC = -2.00 | q.value = 3.53e-08 |