Distributions and lists of all significant differential expression values of the type: SilentIntergenicRegion for comparisons: Pre_vs_Post (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Distribution of log2 differential expression values for comparison: Pre_vs_Post and data type: SilentIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: SilentIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
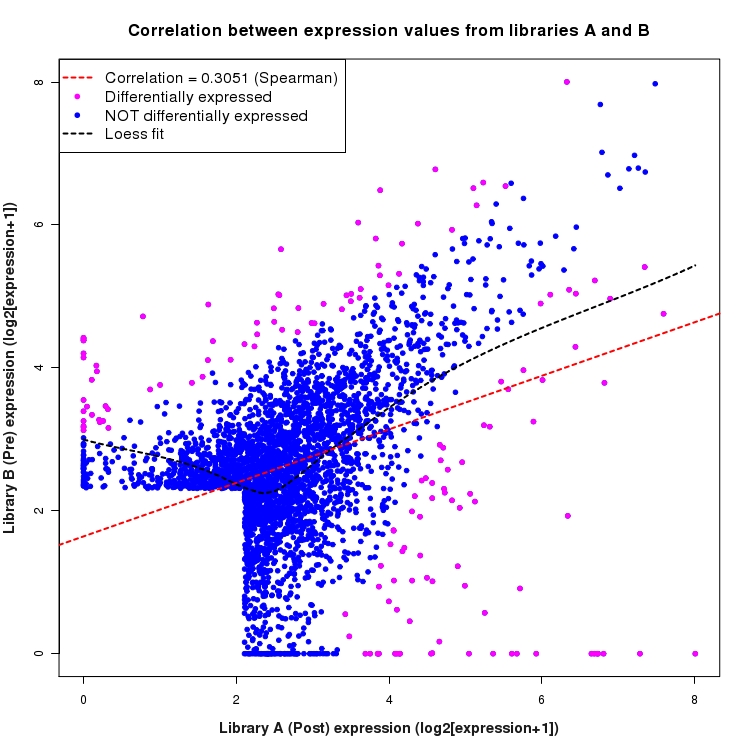
Scatter plot of log2 expression values for comparison: Pre_vs_Post and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
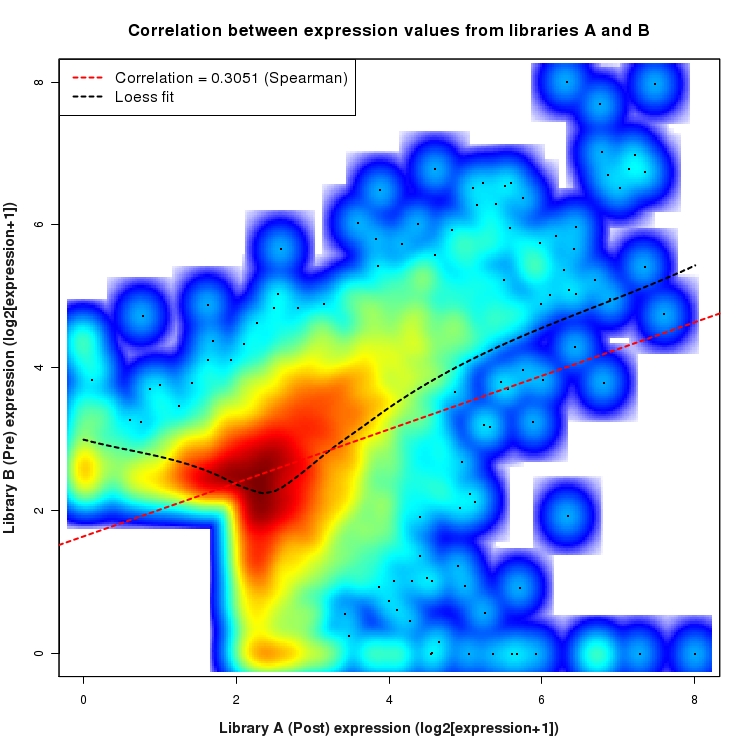
SmoothScatter plot of log2 expression values for comparison: Pre_vs_Post and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed SilentIntergenicRegion features
The following table provides a ranked list of all significant differentially expressed SilentIntergenicRegion features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | Pre_vs_Post (Gene | Feature | Links | Values) |
| 1 | IGLV3-13 | IG1_SR3 | (Pre) | (Post) | FC = 257.72 | q.value = 1.01e-75 |
| 2 | IGLV3-13 | IG1_SR2 | (Pre) | (Post) | FC = 156.00 | q.value = 6.37e-45 |
| 3 | IGLV3-13 | IG1_SR1 | (Pre) | (Post) | FC = 112.09 | q.value = 1.51e-31 |
| 4 | AL122127.6 AL122127.6 | IG23_SR2 | (Pre) | (Post) | FC = 106.69 | q.value = 3.95e-30 |
| 5 | AL122127.6 AL122127.6 | IG23_SR6 | (Pre) | (Post) | FC = 105.73 | q.value = 6.23e-30 |
| 6 | IGLV3-13 | IG1_SR4 | (Pre) | (Post) | FC = 103.11 | q.value = 3.69e-29 |
| 7 | AL122127.6 AL122127.6 | IG23_SR5 | (Pre) | (Post) | FC = 100.41 | q.value = 2.3e-28 |
| 8 | AL122127.6 AL122127.6 | IG23_SR1 | (Pre) | (Post) | FC = 60.85 | q.value = 2.21e-16 |
| 9 | AL122127.6 AL122127.6 | IG23_SR3 | (Pre) | (Post) | FC = 51.03 | q.value = 9.98e-14 |
| 10 | AL122127.6 AL122127.6 | IG23_SR4 | (Pre) | (Post) | FC = 48.76 | q.value = 6.09e-13 |
| 11 | AL122127.6 AL122127.6 | IG23_SR7 | (Pre) | (Post) | FC = 41.14 | q.value = 1.61e-10 |
| 12 | IGLV2-11 IGL@ | IG5_SR1 | (Pre) | (Post) | FC = 33.05 | q.value = 2.18e-08 |
| 13 | D87023.1 D87023.1 | IG20_SR1 | (Pre) | (Post) | FC = 27.90 | q.value = 1.88e-12 |
| 14 | D87023.1 D87023.1 | IG20_SR4 | (Pre) | (Post) | FC = 25.67 | q.value = 1.1e-09 |
| 15 | D87023.1 D87023.1 | IG20_SR6 | (Pre) | (Post) | FC = 23.57 | q.value = 1.31e-05 |
| 16 | IGLV3-25 LL22NC03-102D1.16 | IG38_SR2 | (Pre) | (Post) | FC = 23.43 | q.value = 1.31e-05 |
| 17 | RP11-534K14.2 RNF17 | IG45_SR1 | (Pre) | (Post) | FC = 22.47 | q.value = 4.45e-06 |
| 18 | SREBF2 TNFRSF13C | IG36_SR6 | (Pre) | (Post) | FC = -21.43 | q.value = 1.18e-05 |
| 19 | SNORA76 TEX2 | IG15_SR1 | (Pre) | (Post) | FC = 21.28 | q.value = 2.4e-18 |
| 20 | NAT12 C14orf105 | IG7_SR8 | (Pre) | (Post) | FC = -21.22 | q.value = 1.18e-05 |
| 21 | NAT12 C14orf105 | IG7_SR7 | (Pre) | (Post) | FC = -20.95 | q.value = 1.18e-05 |
| 22 | BHLHE22 CYP7B1 | IG34_SR9 | (Pre) | (Post) | FC = -20.85 | q.value = 1.18e-05 |
| 23 | BHLHE22 CYP7B1 | IG34_SR7 | (Pre) | (Post) | FC = -18.41 | q.value = 0.000102 |
| 24 | IGHVII-31-1 | IG1_SR1 | (Pre) | (Post) | FC = 17.69 | q.value = 0.000797 |
| 25 | GRAMD1B SCN3B | IG27_SR3 | (Pre) | (Post) | FC = -17.68 | q.value = 0.000102 |
| 26 | SNORA24 PRSS12 | IG3_SR2 | (Pre) | (Post) | FC = 17.56 | q.value = 0.000797 |
| 27 | FTL GYS1 | IG32_SR1 | (Pre) | (Post) | FC = 17.17 | q.value = 0.000797 |
| 28 | IGLV3-25 LL22NC03-102D1.16 | IG38_SR1 | (Pre) | (Post) | FC = 16.90 | q.value = 0.00147 |
| 29 | IGHV1-68 IGHV1-69 | IG14_SR3 | (Pre) | (Post) | FC = 16.48 | q.value = 9.73e-07 |
| 30 | AL356017.3 CLMN | IG4_SR3 | (Pre) | (Post) | FC = -15.37 | q.value = 4.06e-06 |
| 31 | IGLV3-19 IGLV2-18 | IG46_SR3 | (Pre) | (Post) | FC = 14.60 | q.value = 0.00245 |
| 32 | KLHDC7B C22orf41 | IG3_SR1 | (Pre) | (Post) | FC = -14.56 | q.value = 0.000401 |
| 33 | NEB ARL5A | IG31_SR9 | (Pre) | (Post) | FC = 14.47 | q.value = 0.00245 |
| 34 | IGHJ2P AL122127.6 | IG25_SR9 | (Pre) | (Post) | FC = 14.43 | q.value = 0.00245 |
| 35 | D87023.1 D87023.1 | IG20_SR3 | (Pre) | (Post) | FC = 14.10 | q.value = 0.000259 |
| 36 | HOXB2 HOXB3 | IG37_SR3 | (Pre) | (Post) | FC = -13.70 | q.value = 0.000797 |
| 37 | FTL GYS1 | IG32_SR3 | (Pre) | (Post) | FC = 13.48 | q.value = 0.00734 |
| 38 | GRAMD1B SCN3B | IG27_SR2 | (Pre) | (Post) | FC = -13.22 | q.value = 0.00159 |
| 39 | IRF2 CASP3 | IG5_SR13 | (Pre) | (Post) | FC = 12.91 | q.value = 0.00734 |
| 40 | RP11-269F19.7 U5 | IG26_SR2 | (Pre) | (Post) | FC = 12.78 | q.value = 5.11e-06 |
| 41 | hsa-mir-29a hsa-mir-29b-1 | IG12_SR1 | (Pre) | (Post) | FC = 11.76 | q.value = 0.000128 |
| 42 | RP11-252C24.2 CTNNA3 | IG16_SR22 | (Pre) | (Post) | FC = -11.71 | q.value = 0.00632 |
| 43 | PPM1L B3GALNT1 | IG20_SR1 | (Pre) | (Post) | FC = 11.22 | q.value = 0.00494 |
| 44 | AL138963.20 AL138963.20 | IG16_SR3 | (Pre) | (Post) | FC = 10.82 | q.value = 0.00022 |
| 45 | LZTFL1 CCR9 | IG41_SR3 | (Pre) | (Post) | FC = -10.64 | q.value = 0.0115 |
| 46 | AC015574.8 AC016251.9 | IG10_SR38 | (Pre) | (Post) | FC = -10.50 | q.value = 0.0115 |
| 47 | AL138963.20 AL138963.20 | IG16_SR2 | (Pre) | (Post) | FC = 9.71 | q.value = 0.001 |
| 48 | CRB1 AL139136.17 | IG6_SR4 | (Pre) | (Post) | FC = 9.62 | q.value = 0.0132 |
| 49 | SUCNR1 MBNL1 | IG16_SR2 | (Pre) | (Post) | FC = -9.56 | q.value = 0.0213 |
| 50 | RP11-389O22.3 RP11-389O22.4 | IG41_SR2 | (Pre) | (Post) | FC = -9.53 | q.value = 2.48e-06 |
| 51 | MTMR2 MAML2 | IG37_SR2 | (Pre) | (Post) | FC = 9.44 | q.value = 0.0198 |
| 52 | SUCNR1 MBNL1 | IG16_SR1 | (Pre) | (Post) | FC = -9.39 | q.value = 0.0213 |
| 53 | CYP7B1 AC060775.5 | IG35_SR3 | (Pre) | (Post) | FC = -9.09 | q.value = 0.0401 |
| 54 | PTPRG C3orf14 | IG36_SR1 | (Pre) | (Post) | FC = -9.04 | q.value = 0.0115 |
| 55 | U8 GRAMD1B | IG26_SR9 | (Pre) | (Post) | FC = -8.99 | q.value = 0.0401 |
| 56 | PKLR FDPS | IG9_SR3 | (Pre) | (Post) | FC = -8.73 | q.value = 0.0401 |
| 57 | IRX3 IRX5 | IG21_SR21 | (Pre) | (Post) | FC = -8.59 | q.value = 0.0115 |
| 58 | FBXW7 TMEM154 | IG48_SR8 | (Pre) | (Post) | FC = -8.43 | q.value = 1.47e-09 |
| 59 | TMEM173 Y_RNA | IG2_SR2 | (Pre) | (Post) | FC = -8.39 | q.value = 0.0213 |
| 60 | CRB1 AL139136.17 | IG6_SR5 | (Pre) | (Post) | FC = 8.23 | q.value = 0.0083 |
| 61 | AP002852.3 AP002852.3 | IG19_SR3 | (Pre) | (Post) | FC = 8.20 | q.value = 0.00263 |
| 62 | DOK6 CD226 | IG34_SR9 | (Pre) | (Post) | FC = 8.17 | q.value = 1.06e-18 |
| 63 | SRP_euk_arch AC007384.3 | IG50_SR2 | (Pre) | (Post) | FC = -8.15 | q.value = 0.0401 |
| 64 | RP11-252C24.2 CTNNA3 | IG16_SR21 | (Pre) | (Post) | FC = -8.00 | q.value = 0.0213 |
| 65 | NEB ARL5A | IG31_SR12 | (Pre) | (Post) | FC = 7.98 | q.value = 5.89e-06 |
| 66 | FTL GYS1 | IG32_SR2 | (Pre) | (Post) | FC = 7.62 | q.value = 0.0213 |
| 67 | IRS1 RHBDD1 | IG22_SR7 | (Pre) | (Post) | FC = 7.38 | q.value = 7.62e-05 |
| 68 | ENPP6 AC079080.5 | IG50_SR6 | (Pre) | (Post) | FC = 7.33 | q.value = 0.0314 |
| 69 | DOK6 CD226 | IG34_SR10 | (Pre) | (Post) | FC = 7.15 | q.value = 3.46e-30 |
| 70 | ZNF518B CLNK | IG29_SR4 | (Pre) | (Post) | FC = -7.13 | q.value = 0.0401 |
| 71 | RFX3 GLIS3 | IG28_SR18 | (Pre) | (Post) | FC = -7.09 | q.value = 0.0236 |
| 72 | hsa-mir-29b-1 U6 | IG13_SR1 | (Pre) | (Post) | FC = 7.09 | q.value = 6.74e-05 |
| 73 | ETNK1 SOX5 | IG18_SR44 | (Pre) | (Post) | FC = -6.77 | q.value = 0.0138 |
| 74 | EIF2AK4 SRP14 | IG10_SR2 | (Pre) | (Post) | FC = 6.69 | q.value = 0.0115 |
| 75 | DNAJB13 UCP2 | IG30_SR2 | (Pre) | (Post) | FC = 6.58 | q.value = 0.0115 |
| 76 | FBXW7 TMEM154 | IG48_SR11 | (Pre) | (Post) | FC = -6.41 | q.value = 0.00251 |
| 77 | RP3-416J7.1 AL035696.14 | IG4_SR2 | (Pre) | (Post) | FC = 6.40 | q.value = 0.000207 |
| 78 | C1orf186 CTSE | IG32_SR2 | (Pre) | (Post) | FC = 6.34 | q.value = 0.0213 |
| 79 | IGHJ3P AL122127.6 | IG21_SR3 | (Pre) | (Post) | FC = 6.25 | q.value = 1.41e-08 |
| 80 | SREBF2 TNFRSF13C | IG36_SR5 | (Pre) | (Post) | FC = -6.07 | q.value = 2.42e-14 |
| 81 | hsa-mir-21 U6 | IG17_SR4 | (Pre) | (Post) | FC = 5.62 | q.value = 0.00932 |
| 82 | AC063924.17 ARID2 | IG35_SR9 | (Pre) | (Post) | FC = 5.61 | q.value = 0.0257 |
| 83 | ZNF518B CLNK | IG29_SR5 | (Pre) | (Post) | FC = -5.58 | q.value = 0.00676 |
| 84 | RFX3 GLIS3 | IG28_SR17 | (Pre) | (Post) | FC = -5.57 | q.value = 0.000101 |
| 85 | MYO1G SNORA9 | IG49_SR2 | (Pre) | (Post) | FC = 5.56 | q.value = 0.00251 |
| 86 | FBXW7 TMEM154 | IG48_SR10 | (Pre) | (Post) | FC = -5.47 | q.value = 0.000168 |
| 87 | ARNTL2 C12orf70 | IG44_SR1 | (Pre) | (Post) | FC = -5.41 | q.value = 2.15e-09 |
| 88 | AC011414.4 TIMD4 | IG28_SR3 | (Pre) | (Post) | FC = 5.33 | q.value = 0.00251 |
| 89 | DOK6 CD226 | IG34_SR8 | (Pre) | (Post) | FC = 5.24 | q.value = 0.00997 |
| 90 | FAM83F | IG1_SR3 | (Pre) | (Post) | FC = -5.17 | q.value = 0.0314 |
| 91 | FAM83F | IG1_SR1 | (Pre) | (Post) | FC = -5.14 | q.value = 0.0011 |
| 92 | CD200 BTLA | IG3_SR7 | (Pre) | (Post) | FC = -5.07 | q.value = 0.000256 |
| 93 | hsa-mir-15a DLEU1 | IG25_SR6 | (Pre) | (Post) | FC = 5.06 | q.value = 0.0257 |
| 94 | GPR160 PHC3 | IG16_SR1 | (Pre) | (Post) | FC = 5.03 | q.value = 0.0257 |
| 95 | SLCO5A1 AC079089.8 | IG16_SR3 | (Pre) | (Post) | FC = -4.98 | q.value = 0.0193 |
| 96 | C18orf58 AP001120.5 | IG9_SR9 | (Pre) | (Post) | FC = 4.96 | q.value = 0.0134 |
| 97 | hsa-mir-21 U6 | IG17_SR1 | (Pre) | (Post) | FC = 4.85 | q.value = 0.000518 |
| 98 | CIITA DEXI | IG28_SR1 | (Pre) | (Post) | FC = -4.67 | q.value = 0.0134 |
| 99 | CDR2 AC106788.3 | IG41_SR9 | (Pre) | (Post) | FC = -4.58 | q.value = 0.00526 |
| 100 | STK17B HECW2 | IG26_SR6 | (Pre) | (Post) | FC = 4.57 | q.value = 0.00457 |
| 101 | CIITA DEXI | IG28_SR3 | (Pre) | (Post) | FC = -4.56 | q.value = 0.0208 |
| 102 | IGHJ3P AL122127.6 | IG21_SR2 | (Pre) | (Post) | FC = 4.53 | q.value = 3.38e-07 |
| 103 | GMPS AC067721.22 | IG37_SR1 | (Pre) | (Post) | FC = 4.52 | q.value = 0.00997 |
| 104 | T U6 | IG20_SR2 | (Pre) | (Post) | FC = -4.50 | q.value = 5.35e-14 |
| 105 | U4 | IG1_SR4 | (Pre) | (Post) | FC = -4.43 | q.value = 0.00213 |
| 106 | hsa-mir-29b-1 U6 | IG13_SR3 | (Pre) | (Post) | FC = 4.43 | q.value = 2.98e-09 |
| 107 | hsa-mir-29b-1 U6 | IG13_SR2 | (Pre) | (Post) | FC = 4.41 | q.value = 0.000278 |
| 108 | TRPM1 KLF13 | IG5_SR8 | (Pre) | (Post) | FC = 4.38 | q.value = 0.0498 |
| 109 | AL022316.2 SERHL | IG2_SR4 | (Pre) | (Post) | FC = -4.18 | q.value = 0.0134 |
| 110 | KCNRG hsa-mir-16-1 | IG23_SR11 | (Pre) | (Post) | FC = 4.13 | q.value = 0.000413 |
| 111 | NEB ARL5A | IG31_SR6 | (Pre) | (Post) | FC = 4.07 | q.value = 0.0143 |
| 112 | RP11-154C3.1 RP11-154C3.3 | IG24_SR3 | (Pre) | (Post) | FC = -4.07 | q.value = 0.00212 |
| 113 | hsa-mir-568 ZBTB20 | IG27_SR4 | (Pre) | (Post) | FC = 4.01 | q.value = 0.0215 |
| 114 | PIK3R6 PIK3R5 | IG16_SR2 | (Pre) | (Post) | FC = -3.95 | q.value = 2.27e-06 |
| 115 | IKZF2 AC079610.4 | IG15_SR2 | (Pre) | (Post) | FC = 3.92 | q.value = 0.0215 |
| 116 | AL122127.6 IGHJ3P | IG20_SR1 | (Pre) | (Post) | FC = 3.83 | q.value = 1.11e-15 |
| 117 | CTH AL354872.9 | IG16_SR2 | (Pre) | (Post) | FC = -3.82 | q.value = 0.0134 |
| 118 | AL122127.6 IGHJ3P | IG20_SR2 | (Pre) | (Post) | FC = 3.80 | q.value = 1.66e-11 |
| 119 | IGHJ3P AL122127.6 | IG21_SR1 | (Pre) | (Post) | FC = 3.63 | q.value = 0.000401 |
| 120 | HEXIM1 HEXIM2 | IG28_SR3 | (Pre) | (Post) | FC = 3.55 | q.value = 0.0148 |
| 121 | ZNRF2 NOD1 | IG38_SR1 | (Pre) | (Post) | FC = 3.46 | q.value = 0.000248 |
| 122 | RP11-255J3.3 RFX3 | IG27_SR21 | (Pre) | (Post) | FC = -3.37 | q.value = 0.00752 |
| 123 | KCNRG hsa-mir-16-1 | IG23_SR12 | (Pre) | (Post) | FC = 3.34 | q.value = 0.043 |
| 124 | TMCC3 AC117494.3 | IG19_SR5 | (Pre) | (Post) | FC = -3.24 | q.value = 0.0277 |
| 125 | COPS4 PLAC8 | IG19_SR3 | (Pre) | (Post) | FC = -3.19 | q.value = 1.84e-23 |
| 126 | AC114776.3 BUB1 | IG6_SR3 | (Pre) | (Post) | FC = 3.16 | q.value = 0.00245 |
| 127 | COL19A1 COL9A1 | IG48_SR2 | (Pre) | (Post) | FC = -3.12 | q.value = 0.0251 |
| 128 | MRPL17 GVIN1 | IG36_SR6 | (Pre) | (Post) | FC = -3.12 | q.value = 1.47e-05 |
| 129 | AC009053.7 GLG1 | IG39_SR8 | (Pre) | (Post) | FC = -3.03 | q.value = 0.0251 |
| 130 | hsa-mir-1205 U1 | IG50_SR9 | (Pre) | (Post) | FC = -2.97 | q.value = 0.00906 |
| 131 | CHST7 SLC9A7 | IG12_SR2 | (Pre) | (Post) | FC = -2.97 | q.value = 0.00216 |
| 132 | LNPEP AC008865.3 | IG45_SR1 | (Pre) | (Post) | FC = -2.96 | q.value = 0.000397 |
| 133 | U4 | IG1_SR5 | (Pre) | (Post) | FC = -2.89 | q.value = 0.0115 |
| 134 | CHST7 SLC9A7 | IG12_SR3 | (Pre) | (Post) | FC = -2.78 | q.value = 0.014 |
| 135 | AL122127.6 IGHJ3P | IG20_SR3 | (Pre) | (Post) | FC = 2.77 | q.value = 1.47e-06 |
| 136 | INSL3 JAK3 | IG50_SR2 | (Pre) | (Post) | FC = -2.71 | q.value = 0.0392 |
| 137 | hsa-mir-29b-1 U6 | IG13_SR38 | (Pre) | (Post) | FC = -2.70 | q.value = 0.0228 |
| 138 | hsa-mir-1205 U1 | IG50_SR8 | (Pre) | (Post) | FC = -2.66 | q.value = 0.00984 |
| 139 | AGGF1 ZBED3 | IG33_SR5 | (Pre) | (Post) | FC = -2.66 | q.value = 3.66e-06 |
| 140 | FAM38B2 C18orf58 | IG8_SR2 | (Pre) | (Post) | FC = 2.65 | q.value = 4.52e-05 |
| 141 | PRMT8 AC005831.1 | IG32_SR4 | (Pre) | (Post) | FC = -2.58 | q.value = 0.0277 |
| 142 | AC097484.3 PPM1K | IG11_SR3 | (Pre) | (Post) | FC = -2.57 | q.value = 1.61e-06 |
| 143 | GLS STAT1 | IG12_SR4 | (Pre) | (Post) | FC = 2.40 | q.value = 0.000354 |
| 144 | GVIN1 OR2AG2 | IG37_SR1 | (Pre) | (Post) | FC = -2.27 | q.value = 0.0301 |
| 145 | CHST7 SLC9A7 | IG12_SR5 | (Pre) | (Post) | FC = -2.24 | q.value = 0.0418 |
| 146 | LRAP LNPEP | IG44_SR1 | (Pre) | (Post) | FC = -2.19 | q.value = 0.00097 |
| 147 | LARS2 LIMD1 | IG37_SR6 | (Pre) | (Post) | FC = -2.15 | q.value = 0.00726 |
| 148 | U6 AC010455.5 | IG20_SR5 | (Pre) | (Post) | FC = 2.12 | q.value = 0.0115 |
| 149 | GLS STAT1 | IG12_SR3 | (Pre) | (Post) | FC = 2.12 | q.value = 0.0166 |
| 150 | TCF12 CGNL1 | IG34_SR7 | (Pre) | (Post) | FC = -2.03 | q.value = 0.000557 |