Distributions and lists of all significant differential expression values of the type: Intergenic for comparisons: PostTx_vs_PreTx_Un, PostTx_vs_PreTx_En, En_vs_Un_PreTx, En_vs_Un_PostTx (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Distribution of log2 differential expression values for comparison: PostTx_vs_PreTx_Un and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
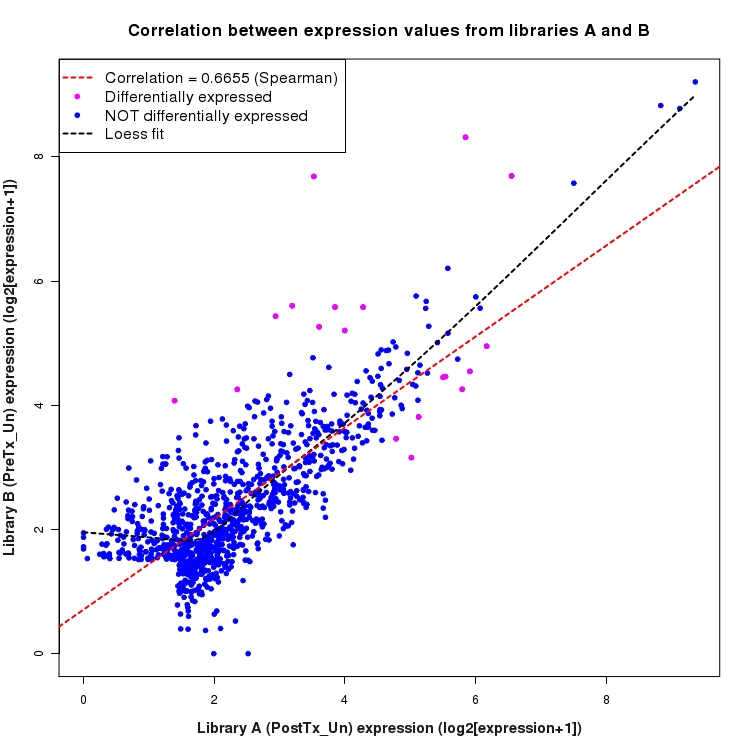
Scatter plot of log2 expression values for comparison: PostTx_vs_PreTx_Un and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
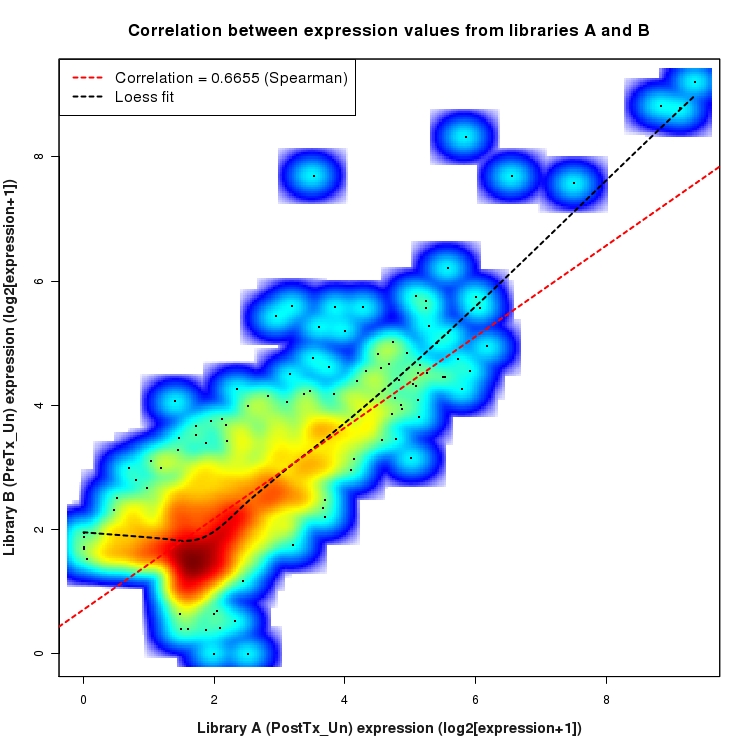
SmoothScatter plot of log2 expression values for comparison: PostTx_vs_PreTx_Un and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Distribution of log2 differential expression values for comparison: PostTx_vs_PreTx_En and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
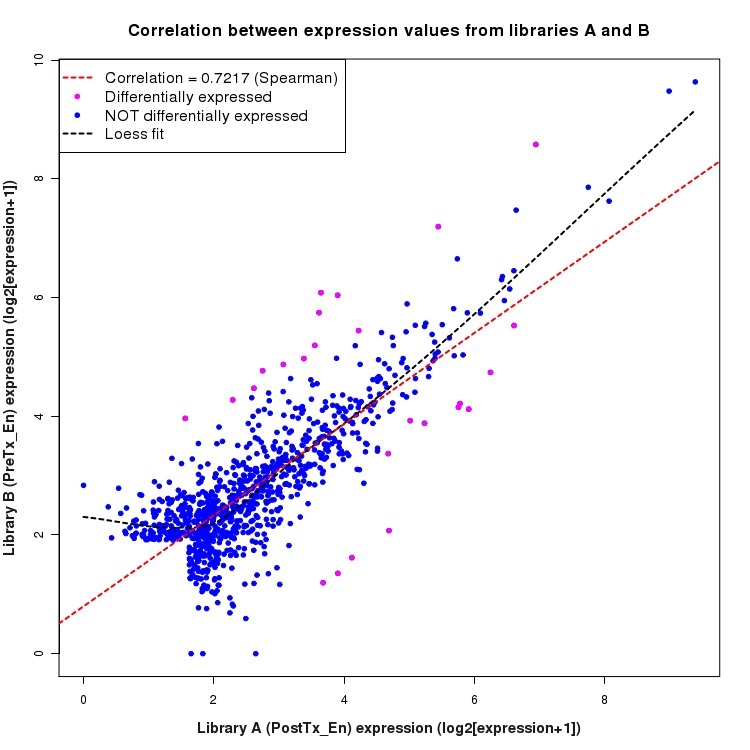
Scatter plot of log2 expression values for comparison: PostTx_vs_PreTx_En and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
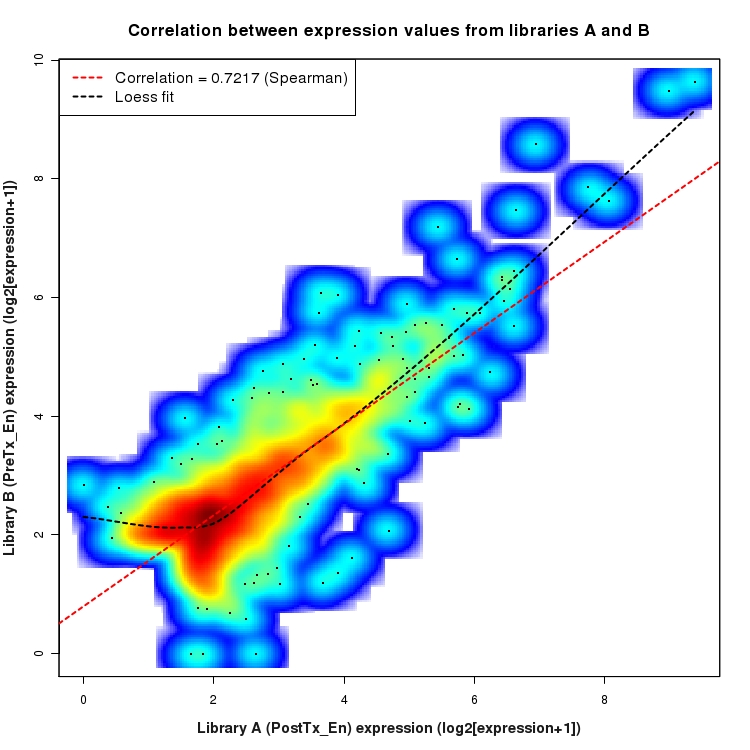
SmoothScatter plot of log2 expression values for comparison: PostTx_vs_PreTx_En and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Distribution of log2 differential expression values for comparison: En_vs_Un_PreTx and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
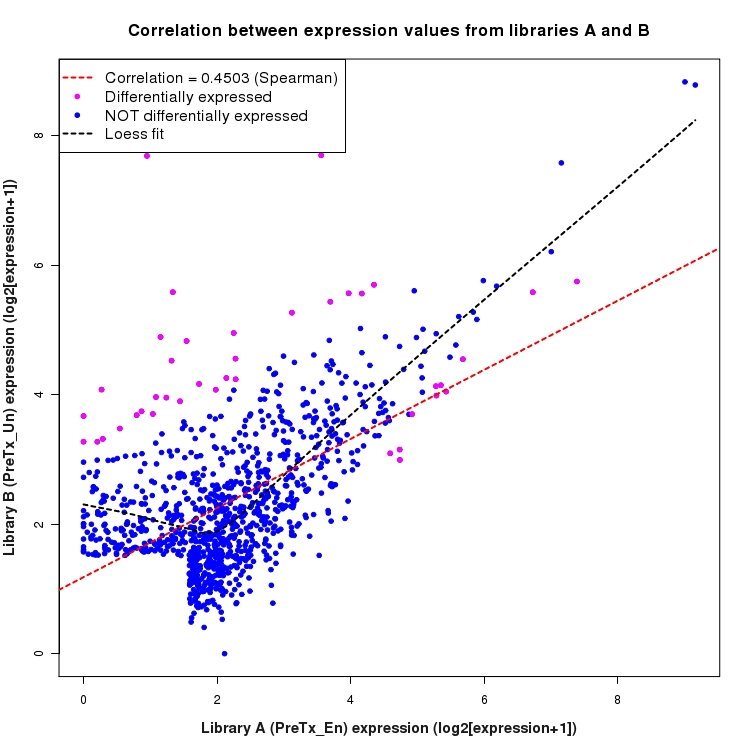
Scatter plot of log2 expression values for comparison: En_vs_Un_PreTx and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
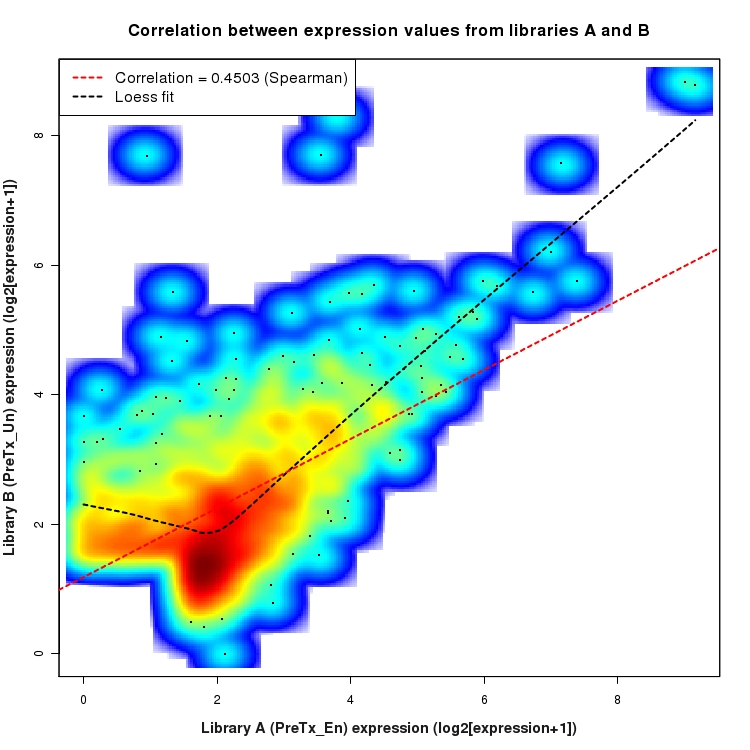
SmoothScatter plot of log2 expression values for comparison: En_vs_Un_PreTx and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Distribution of log2 differential expression values for comparison: En_vs_Un_PostTx and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
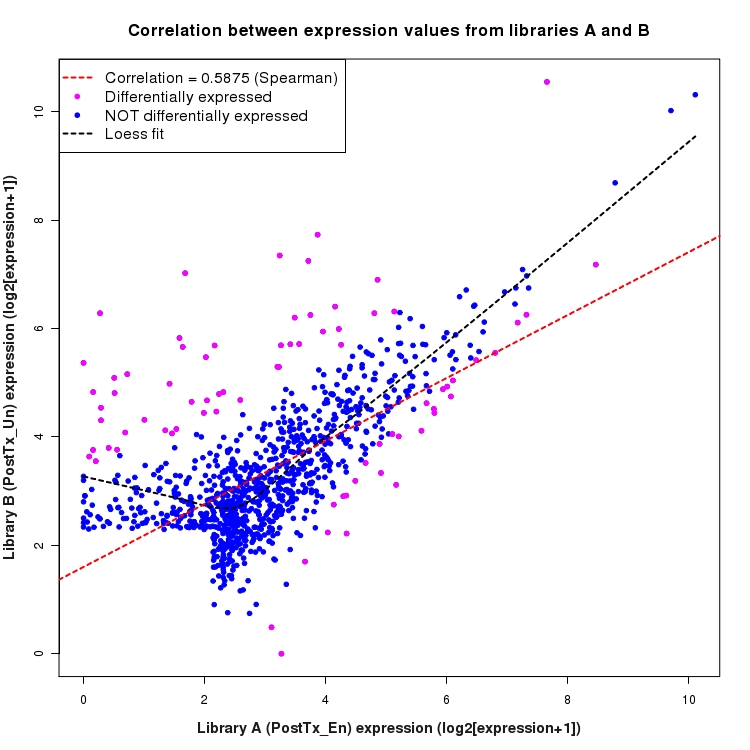
Scatter plot of log2 expression values for comparison: En_vs_Un_PostTx and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
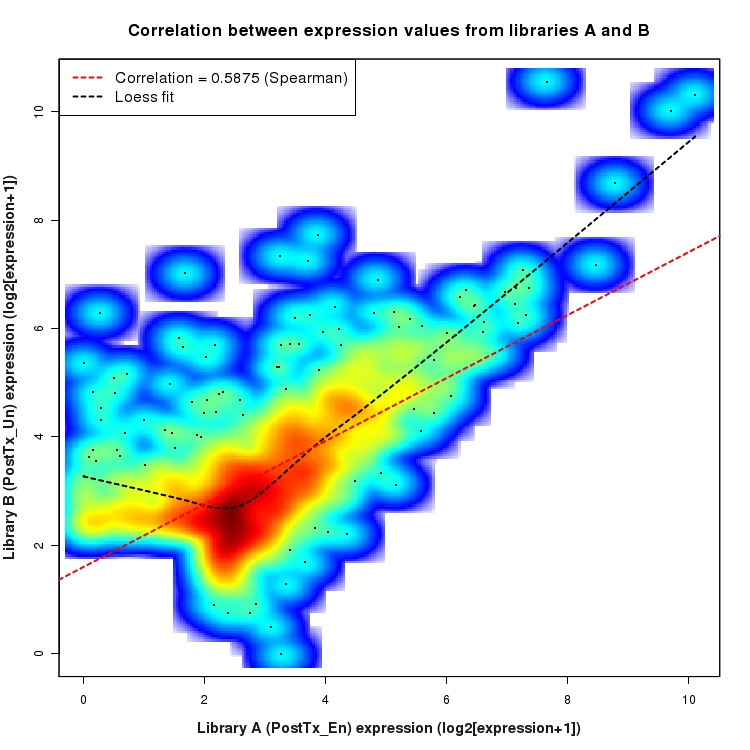
SmoothScatter plot of log2 expression values for comparison: En_vs_Un_PostTx and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed Intergenic features
The following table provides a ranked list of all significant differentially expressed Intergenic features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | PostTx_vs_PreTx_Un (Gene | Feature | Links | Values) | PostTx_vs_PreTx_En (Gene | Feature | Links | Values) | En_vs_Un_PreTx (Gene | Feature | Links | Values) | En_vs_Un_PostTx (Gene | Feature | Links | Values) |
| 1 | SSU_rRNA_5 | IG4 | PostTx_Un | PreTx_Un | FC = -17.89 | q.value = 1.95e-76 | SNORND104 SNORA76 | IG14 | (PostTx_En) | (PreTx_En) | FC = 6.12 | q.value = 4.67e-05 | SSU_rRNA_5 | IG4 | PreTx_En | PreTx_Un | FC = -106.45 | q.value = 7.48e-70 | hsa-mir-223 VSIG4 | IG3 | (PostTx_En) | (PostTx_Un) | FC = -64.39 | q.value = 1.1e-18 |
| 2 | HCFC1 | IG51 | (PostTx_Un) | (PreTx_Un) | FC = -6.42 | q.value = 0.000416 | DULLARD C17orf81 | IG50 | (PostTx_En) | (PreTx_En) | FC = 5.86 | q.value = 0.017 | EPX MKS1 | IG42 | (PreTx_En) | (PreTx_Un) | FC = -23.28 | q.value = 7.53e-88 | hsa-mir-451 hsa-mir-144 | IG47 | (PostTx_En) | (PostTx_Un) | FC = -41.22 | q.value = 3.87e-09 |
| 3 | H1FX C3orf47 | IG7 | (PostTx_Un) | (PreTx_Un) | FC = -5.64 | q.value = 1.04e-08 | RAB14 | IG51 | (PostTx_En) | (PreTx_En) | FC = 5.65 | q.value = 0.00452 | ARID3B CLK3 | IG41 | (PreTx_En) | (PreTx_Un) | FC = -18.93 | q.value = 1.16e-12 | EPX MKS1 | IG42 | (PostTx_En) | (PostTx_Un) | FC = -40.52 | q.value = 7.69e-29 |
| 4 | EPX MKS1 | IG42 | (PostTx_Un) | (PreTx_Un) | FC = -5.54 | q.value = 1.85e-67 | SSU_rRNA_5 | IG4 | PostTx_En | PreTx_En | FC = 5.58 | q.value = 0.0316 | SLC25A37 AC051642.5 | IG28 | (PreTx_En) | (PreTx_Un) | FC = -17.54 | q.value = 3.02e-53 | CLEC4D CLEC4E | IG32 | (PostTx_En) | (PostTx_Un) | FC = -25.42 | q.value = 2.76e-06 |
| 5 | FAM38A AC138028.1 | IG6 | (PostTx_Un) | (PreTx_Un) | FC = -5.31 | q.value = 9.05e-10 | hsa-mir-23a AC020916.8 | IG38 | (PostTx_En) | (PreTx_En) | FC = -5.42 | q.value = 1.23e-09 | LPO MPO | IG44 | (PreTx_En) | (PreTx_Un) | FC = -13.99 | q.value = 0.000159 | TRIM14 CORO2A | IG11 | (PostTx_En) | (PostTx_Un) | FC = -23.93 | q.value = 1.59e-07 |
| 6 | CSNK1G2 BTBD2 | IG29 | (PostTx_Un) | (PreTx_Un) | FC = -3.74 | q.value = 0.0136 | GNAL MPPE1 | IG11 | (PostTx_En) | (PreTx_En) | FC = -5.29 | q.value = 0.0168 | CEBPD | IG1 | (PreTx_En) | (PreTx_Un) | FC = -13.32 | q.value = 9.4e-07 | AL137792.11 PTAFR | IG12 | (PostTx_En) | (PostTx_Un) | FC = -21.62 | q.value = 1.01e-06 |
| 7 | U3 U3 | IG41 | (PostTx_Un) | (PreTx_Un) | FC = 3.63 | q.value = 0.000618 | AC116366.4 IRF1 | IG14 | (PostTx_En) | (PreTx_En) | FC = -4.41 | q.value = 1.22e-07 | hsa-mir-451 hsa-mir-144 | IG47 | (PreTx_En) | (PreTx_Un) | FC = -12.72 | q.value = 0.00303 | ERAL1 hsa-mir-451 | IG46 | (PostTx_En) | (PostTx_Un) | FC = -19.63 | q.value = 4.53e-06 |
| 8 | ARID3B CLK3 | IG41 | (PostTx_Un) | (PreTx_Un) | FC = -3.33 | q.value = 1.3e-05 | SLC35A2 PIM2 | IG23 | (PostTx_En) | (PreTx_En) | FC = -4.38 | q.value = 2.6e-06 | EXOSC9 CCNA2 | IG29 | (PreTx_En) | (PreTx_Un) | FC = -9.73 | q.value = 2.23e-05 | LPO MPO | IG44 | (PostTx_En) | (PostTx_Un) | FC = -19.00 | q.value = 6.42e-05 |
| 9 | SNORD78 SNORD44 | IG49 | (PostTx_Un) | (PreTx_Un) | FC = -3.16 | q.value = 0.000586 | hsa-mir-27a hsa-mir-23a | IG37 | (PostTx_En) | (PreTx_En) | FC = -4.06 | q.value = 0.00479 | COX4NB COX4I1 | IG27 | (PreTx_En) | (PreTx_Un) | FC = -9.66 | q.value = 0.0164 | CEBPD | IG1 | (PostTx_En) | (PostTx_Un) | FC = -18.88 | q.value = 9.94e-11 |
| 10 | PLXNC1 CCDC41 | IG14 | (PostTx_Un) | (PreTx_Un) | FC = 2.90 | q.value = 2.23e-05 | hsa-mir-221 hsa-mir-222 | IG50 | (PostTx_En) | (PreTx_En) | FC = -3.96 | q.value = 0.0395 | RAB14 | IG51 | (PreTx_En) | (PreTx_Un) | FC = -9.22 | q.value = 0.000168 | DTX4 MPEG1 | IG22 | (PostTx_En) | (PostTx_Un) | FC = -17.21 | q.value = 2.93e-30 |
| 11 | CFLAR AC007283.3 | IG15 | (PostTx_Un) | (PreTx_Un) | FC = 2.58 | q.value = 4.93e-05 | AC020659.5 EHD4 | IG8 | (PostTx_En) | (PreTx_En) | FC = -3.62 | q.value = 0.0339 | TRIM14 CORO2A | IG11 | (PreTx_En) | (PreTx_Un) | FC = -8.37 | q.value = 0.0164 | EXOSC9 CCNA2 | IG29 | (PostTx_En) | (PostTx_Un) | FC = -16.23 | q.value = 4.31e-09 |
| 12 | C2orf29 SNORD89 | IG43 | (PostTx_Un) | (PreTx_Un) | FC = 2.50 | q.value = 0.0464 | SRP_euk_arch AL356356.17 | IG27 | (PostTx_En) | (PreTx_En) | FC = -3.50 | q.value = 0.0135 | ERAL1 hsa-mir-451 | IG46 | (PreTx_En) | (PreTx_Un) | FC = -8.15 | q.value = 0.0164 | RP1-229K20.5 TREM1 | IG5 | (PostTx_En) | (PostTx_Un) | FC = -16.20 | q.value = 0.000627 |
| 13 | HIPK3 AL122015.17 | IG30 | (PostTx_Un) | (PreTx_Un) | FC = 2.49 | q.value = 0.0169 | AC100778.5 SNORD58 | IG27 | (PostTx_En) | (PreTx_En) | FC = 3.46 | q.value = 1.47e-07 | SHARPIN MAF1 | IG4 | (PreTx_En) | (PreTx_Un) | FC = -7.63 | q.value = 0.042 | SLC25A37 AC051642.5 | IG28 | (PostTx_En) | (PostTx_Un) | FC = -14.55 | q.value = 3.14e-36 |
| 14 | NP AL355075.6 | IG62 | (PostTx_Un) | (PreTx_Un) | FC = -2.47 | q.value = 0.00216 | NP AL355075.6 | IG62 | (PostTx_En) | (PreTx_En) | FC = -3.37 | q.value = 1.22e-12 | KIAA0329 ANKRD9 | IG4 | (PreTx_En) | (PreTx_Un) | FC = -7.39 | q.value = 0.0128 | ZNF414 MYO1F | IG35 | (PostTx_En) | (PostTx_Un) | FC = -12.12 | q.value = 0.00771 |
| 15 | DTX4 MPEG1 | IG22 | (PostTx_Un) | (PreTx_Un) | FC = 2.32 | q.value = 4.99e-05 | hsa-mir-570 AC069513.28 | IG49 | (PostTx_En) | (PreTx_En) | FC = -3.13 | q.value = 0.00758 | DULLARD C17orf81 | IG50 | (PreTx_En) | (PreTx_Un) | FC = -7.34 | q.value = 0.0104 | ARID3B CLK3 | IG41 | (PostTx_En) | (PostTx_Un) | FC = -11.78 | q.value = 6.58e-05 |
| 16 | hsa-mir-23a AC020916.8 | IG38 | (PostTx_Un) | (PreTx_Un) | FC = -2.31 | q.value = 0.0232 | AC114498.2 AC114498.2 | IG22 | (PostTx_En) | (PreTx_En) | FC = -3.12 | q.value = 1.57e-30 | SNORD60 TRAF7 | IG45 | (PreTx_En) | (PreTx_Un) | FC = -7.32 | q.value = 0.0128 | CST7 C20orf3 | IG3 | (PostTx_En) | (PostTx_Un) | FC = -11.65 | q.value = 0.0224 |
| 17 | SLC25A37 AC051642.5 | IG28 | (PostTx_Un) | (PreTx_Un) | FC = -2.21 | q.value = 2.68e-12 | SNORD81 SNORD47 | IG45 | (PostTx_En) | (PreTx_En) | FC = 3.03 | q.value = 9.71e-06 | RPL36 LONP1 | IG13 | (PreTx_En) | (PreTx_Un) | FC = -6.55 | q.value = 0.015 | COPS4 PLAC8 | IG19 | (PostTx_En) | (PostTx_Un) | FC = -11.56 | q.value = 3.51e-24 |
| 18 | GPR137C ERO1L | IG16 | (PostTx_Un) | (PreTx_Un) | FC = 2.11 | q.value = 0.0161 | ISGF3G REC8 | IG9 | (PostTx_En) | (PreTx_En) | FC = -3.01 | q.value = 0.0407 | DTX4 MPEG1 | IG22 | (PreTx_En) | (PreTx_Un) | FC = -6.50 | q.value = 4.3e-05 | IL17RA CECR6 | IG63 | (PostTx_En) | (PostTx_Un) | FC = -11.47 | q.value = 1.19e-08 |
| 19 | SNORD47 snosnR60_Z15 | IG46 | (PostTx_Un) | (PreTx_Un) | FC = 2.07 | q.value = 0.0246 | NEK6 PSMB7 | IG42 | (PostTx_En) | (PreTx_En) | FC = 2.96 | q.value = 9.71e-06 | AC087749.12 AC087749.12 | IG24 | (PreTx_En) | (PreTx_Un) | FC = -6.33 | q.value = 0.0128 | CCNDBP1 EPB42 | IG27 | (PostTx_En) | (PostTx_Un) | FC = -10.92 | q.value = 4.69e-07 |
| 20 | EMPTY | SNORD47 snosnR60_Z15 | IG46 | (PostTx_En) | (PreTx_En) | FC = 2.84 | q.value = 1.47e-07 | IL17RA CECR6 | IG63 | (PreTx_En) | (PreTx_Un) | FC = -5.46 | q.value = 0.015 | STX3 MRPL16 | IG41 | (PostTx_En) | (PostTx_Un) | FC = -10.48 | q.value = 0.015 |
| 21 | EMPTY | SNORD79 SNORD78 | IG48 | (PostTx_En) | (PreTx_En) | FC = 2.55 | q.value = 0.0045 | SNORND104 SNORA76 | IG14 | (PreTx_En) | (PreTx_Un) | FC = -5.40 | q.value = 0.0128 | SETD8 RILPL2 | IG13 | (PostTx_En) | (PostTx_Un) | FC = -10.41 | q.value = 0.00771 |
| 22 | EMPTY | TRIM44 | IG51 | (PostTx_En) | (PreTx_En) | FC = 2.47 | q.value = 0.0393 | CCNDBP1 EPB42 | IG27 | (PreTx_En) | (PreTx_Un) | FC = -4.85 | q.value = 0.00557 | hsa-mir-373 NALP12 | IG11 | (PostTx_En) | (PostTx_Un) | FC = -10.19 | q.value = 0.0224 |
| 23 | EMPTY | ZSWIM4 hsa-mir-24-2 | IG35 | (PostTx_En) | (PreTx_En) | FC = -2.33 | q.value = 0.048 | SNORD78 SNORD44 | IG49 | (PreTx_En) | (PreTx_Un) | FC = -4.41 | q.value = 0.000245 | SSU_rRNA_5 RNF141 | IG43 | (PostTx_En) | (PostTx_Un) | FC = -9.91 | q.value = 0.00202 |
| 24 | EMPTY | RP11-764K9.3 RP11-764K9.2 | IG33 | (PostTx_En) | (PreTx_En) | FC = 2.12 | q.value = 0.0346 | CSNK1G2 BTBD2 | IG29 | (PreTx_En) | (PreTx_Un) | FC = -4.34 | q.value = 0.0293 | AC004985.2 UBE2D4 | IG25 | (PostTx_En) | (PostTx_Un) | FC = 9.64 | q.value = 0.00673 |
| 25 | EMPTY | PLXNC1 CCDC41 | IG14 | (PostTx_En) | (PreTx_En) | FC = 2.11 | q.value = 2.11e-06 | HCFC1 | IG51 | (PreTx_En) | (PreTx_Un) | FC = -4.27 | q.value = 0.0316 | GRAMD1B SCN3B | IG27 | (PostTx_En) | (PostTx_Un) | FC = -9.23 | q.value = 0.00771 |
| 26 | EMPTY | EMPTY | DOT1L PLEKHJ1 | IG34 | (PreTx_En) | (PreTx_Un) | FC = -3.88 | q.value = 0.0293 | AC114498.2 AC114498.2 | IG22 | (PostTx_En) | (PostTx_Un) | FC = -7.43 | q.value = 6e-200 |
| 27 | EMPTY | EMPTY | hsa-mir-570 AC069513.28 | IG49 | (PreTx_En) | (PreTx_Un) | FC = 3.36 | q.value = 0.00782 | C1orf26 IVNS1ABP | IG46 | (PostTx_En) | (PostTx_Un) | FC = -7.27 | q.value = 0.00122 |
| 28 | EMPTY | EMPTY | H1FX C3orf47 | IG7 | (PreTx_En) | (PreTx_Un) | FC = -3.33 | q.value = 0.00193 | RBKS BRE | IG5 | (PostTx_En) | (PostTx_Un) | FC = -6.82 | q.value = 0.0275 |
| 29 | EMPTY | EMPTY | hsa-mir-29c hsa-mir-29b-2 | IG15 | (PreTx_En) | (PreTx_Un) | FC = 3.13 | q.value = 4.88e-17 | GTF3A MTIF3 | IG24 | (PostTx_En) | (PostTx_Un) | FC = -6.55 | q.value = 1.19e-08 |
| 30 | EMPTY | EMPTY | COPS4 PLAC8 | IG19 | (PreTx_En) | (PreTx_Un) | FC = -3.01 | q.value = 0.00193 | SNORD60 TRAF7 | IG45 | (PostTx_En) | (PostTx_Un) | FC = -6.20 | q.value = 0.00239 |
| 31 | EMPTY | EMPTY | DOPEY1 PGM3 | IG31 | (PreTx_En) | (PreTx_Un) | FC = 3.00 | q.value = 0.0128 | STYXL1 MDH2 | IG2 | (PostTx_En) | (PostTx_Un) | FC = 6.14 | q.value = 0.0143 |
| 32 | EMPTY | EMPTY | FCRL2 FCRL1 | IG18 | (PreTx_En) | (PreTx_Un) | FC = 2.83 | q.value = 0.047 | AL138963.20 TPT1 | IG17 | (PostTx_En) | (PostTx_Un) | FC = -6.11 | q.value = 0.0275 |
| 33 | EMPTY | EMPTY | NACA | IG51 | (PreTx_En) | (PreTx_Un) | FC = -2.62 | q.value = 0.0104 | WDFY2 DHRS12 | IG40 | (PostTx_En) | (PostTx_Un) | FC = -6.07 | q.value = 0.0428 |
| 34 | EMPTY | EMPTY | hsa-mir-29b-2 C1orf132 | IG16 | (PreTx_En) | (PreTx_Un) | FC = 2.62 | q.value = 0.00439 | MARCH7 LY75 | IG27 | (PostTx_En) | (PostTx_Un) | FC = -5.87 | q.value = 0.00419 |
| 35 | EMPTY | EMPTY | ALOX5 MARCH8 | IG13 | (PreTx_En) | (PreTx_Un) | FC = -2.54 | q.value = 0.00896 | AD000864.1 NFKBID | IG49 | (PostTx_En) | (PostTx_Un) | FC = -5.72 | q.value = 0.00267 |
| 36 | EMPTY | EMPTY | SLC35A2 PIM2 | IG23 | (PreTx_En) | (PreTx_Un) | FC = 2.47 | q.value = 0.00896 | SLC18A2 PDZD8 | IG6 | (PostTx_En) | (PostTx_Un) | FC = -5.66 | q.value = 3.96e-08 |
| 37 | EMPTY | EMPTY | RABGAP1L Z99127.2 | IG9 | (PreTx_En) | (PreTx_Un) | FC = 2.34 | q.value = 0.0451 | MCM4 AC021236.10 | IG4 | (PostTx_En) | (PostTx_Un) | FC = -5.48 | q.value = 0.0143 |
| 38 | EMPTY | EMPTY | ENAM IGJ | IG14 | (PreTx_En) | (PreTx_Un) | FC = 2.31 | q.value = 0.0148 | PANK2 RNF24 | IG26 | (PostTx_En) | (PostTx_Un) | FC = -5.38 | q.value = 4.36e-05 |
| 39 | EMPTY | EMPTY | NP AL355075.6 | IG62 | (PreTx_En) | (PreTx_Un) | FC = 2.22 | q.value = 2.43e-06 | AP001266.4 AP001266.4 | IG21 | (PostTx_En) | (PostTx_Un) | FC = -4.89 | q.value = 6.42e-05 |
| 40 | EMPTY | EMPTY | SLAMF7 | IG51 | (PreTx_En) | (PreTx_Un) | FC = 2.22 | q.value = 0.0339 | FAM105A AC010627.7 | IG29 | (PostTx_En) | (PostTx_Un) | FC = -4.83 | q.value = 0.0312 |
| 41 | EMPTY | EMPTY | CFLAR AC007283.3 | IG15 | (PreTx_En) | (PreTx_Un) | FC = 2.20 | q.value = 0.00439 | NACA | IG51 | (PostTx_En) | (PostTx_Un) | FC = -4.74 | q.value = 1.78e-07 |
| 42 | EMPTY | EMPTY | EMPTY | PXK PDHB | IG25 | (PostTx_En) | (PostTx_Un) | FC = -4.44 | q.value = 0.000218 |
| 43 | EMPTY | EMPTY | EMPTY | AL354865.9 | IG51 | (PostTx_En) | (PostTx_Un) | FC = 4.37 | q.value = 0.00245 |
| 44 | EMPTY | EMPTY | EMPTY | C19orf39 EPOR | IG11 | (PostTx_En) | (PostTx_Un) | FC = -4.26 | q.value = 0.0224 |
| 45 | EMPTY | EMPTY | EMPTY | MORF4L1 AC011944.12 | IG7 | (PostTx_En) | (PostTx_Un) | FC = -4.26 | q.value = 0.00151 |
| 46 | EMPTY | EMPTY | EMPTY | TOPORS AL353671.6 | IG14 | (PostTx_En) | (PostTx_Un) | FC = -4.17 | q.value = 0.00213 |
| 47 | EMPTY | EMPTY | EMPTY | XRRA1 SPCS2 | IG47 | (PostTx_En) | (PostTx_Un) | FC = 4.14 | q.value = 8.17e-06 |
| 48 | EMPTY | EMPTY | EMPTY | SNORD89 RNF149 | IG44 | (PostTx_En) | (PostTx_Un) | FC = -4.11 | q.value = 4.31e-09 |
| 49 | EMPTY | EMPTY | EMPTY | C2orf29 SNORD89 | IG43 | (PostTx_En) | (PostTx_Un) | FC = -3.97 | q.value = 0.000184 |
| 50 | EMPTY | EMPTY | EMPTY | C10orf59 | IG51 | (PostTx_En) | (PostTx_Un) | FC = 3.89 | q.value = 0.0206 |
| 51 | EMPTY | EMPTY | EMPTY | C1orf27 | IG1 | (PostTx_En) | (PostTx_Un) | FC = 3.48 | q.value = 0.0223 |
| 52 | EMPTY | EMPTY | EMPTY | PCBP2 MAP3K12 | IG6 | (PostTx_En) | (PostTx_Un) | FC = -3.42 | q.value = 0.000843 |
| 53 | EMPTY | EMPTY | EMPTY | AC013414.7 AC013414.7 | IG30 | (PostTx_En) | (PostTx_Un) | FC = 2.99 | q.value = 0.00132 |
| 54 | EMPTY | EMPTY | EMPTY | RAB14 | IG51 | (PostTx_En) | (PostTx_Un) | FC = -2.78 | q.value = 0.00133 |
| 55 | EMPTY | EMPTY | EMPTY | AL121586.31 FER1L4 | IG33 | (PostTx_En) | (PostTx_Un) | FC = 2.77 | q.value = 5.16e-05 |
| 56 | EMPTY | EMPTY | EMPTY | HNRNPA1 NFE2 | IG26 | (PostTx_En) | (PostTx_Un) | FC = -2.72 | q.value = 0.0224 |
| 57 | EMPTY | EMPTY | EMPTY | AC096887.2 RFT1 | IG32 | (PostTx_En) | (PostTx_Un) | FC = 2.68 | q.value = 0.0253 |
| 58 | EMPTY | EMPTY | EMPTY | LCAT SLC12A4 | IG14 | (PostTx_En) | (PostTx_Un) | FC = 2.61 | q.value = 0.0253 |
| 59 | EMPTY | EMPTY | EMPTY | PSME2 RNF31 | IG8 | (PostTx_En) | (PostTx_Un) | FC = 2.61 | q.value = 0.0297 |
| 60 | EMPTY | EMPTY | EMPTY | TMEM167A XRCC4 | IG37 | (PostTx_En) | (PostTx_Un) | FC = 2.56 | q.value = 2.62e-05 |
| 61 | EMPTY | EMPTY | EMPTY | DEPDC6 | IG51 | (PostTx_En) | (PostTx_Un) | FC = 2.50 | q.value = 3.47e-06 |
| 62 | EMPTY | EMPTY | EMPTY | ABCB8 ACCN3 | IG38 | (PostTx_En) | (PostTx_Un) | FC = 2.46 | q.value = 0.0297 |
| 63 | EMPTY | EMPTY | EMPTY | hsa-mir-29c hsa-mir-29b-2 | IG15 | (PostTx_En) | (PostTx_Un) | FC = 2.44 | q.value = 1.46e-31 |
| 64 | EMPTY | EMPTY | EMPTY | AC013414.7 RTN4 | IG31 | (PostTx_En) | (PostTx_Un) | FC = 2.42 | q.value = 7.02e-05 |
| 65 | EMPTY | EMPTY | EMPTY | SLAMF7 | IG51 | (PostTx_En) | (PostTx_Un) | FC = 2.38 | q.value = 2.73e-09 |
| 66 | EMPTY | EMPTY | EMPTY | ANKRD37 UFSP2 | IG14 | (PostTx_En) | (PostTx_Un) | FC = 2.30 | q.value = 0.00205 |
| 67 | EMPTY | EMPTY | EMPTY | MOBKL1B | IG1 | (PostTx_En) | (PostTx_Un) | FC = -2.26 | q.value = 0.0234 |
| 68 | EMPTY | EMPTY | EMPTY | SNORA67 C6orf105 | IG20 | (PostTx_En) | (PostTx_Un) | FC = 2.20 | q.value = 0.0312 |
| 69 | EMPTY | EMPTY | EMPTY | SNORD27 SNORD26 | IG31 | (PostTx_En) | (PostTx_Un) | FC = 2.11 | q.value = 8.76e-05 |
| 70 | EMPTY | EMPTY | EMPTY | NEK6 PSMB7 | IG42 | (PostTx_En) | (PostTx_Un) | FC = 2.10 | q.value = 1.46e-06 |
| 71 | EMPTY | EMPTY | EMPTY | BHLHA15 AC091654.4 | IG32 | (PostTx_En) | (PostTx_Un) | FC = 2.09 | q.value = 4.68e-11 |
| 72 | EMPTY | EMPTY | EMPTY | SNORD29 SNORD28 | IG29 | (PostTx_En) | (PostTx_Un) | FC = 2.09 | q.value = 6.09e-10 |
| 73 | EMPTY | EMPTY | EMPTY | FCRL2 FCRL1 | IG18 | (PostTx_En) | (PostTx_Un) | FC = 2.09 | q.value = 5.46e-05 |
| 74 | EMPTY | EMPTY | EMPTY | SNORD79 SNORD78 | IG48 | (PostTx_En) | (PostTx_Un) | FC = 2.08 | q.value = 0.000222 |
| 75 | EMPTY | EMPTY | EMPTY | CALML4 CLN6 | IG28 | (PostTx_En) | (PostTx_Un) | FC = 2.07 | q.value = 0.0126 |
| 76 | EMPTY | EMPTY | EMPTY | hsa-mir-29b-2 C1orf132 | IG16 | (PostTx_En) | (PostTx_Un) | FC = 2.06 | q.value = 0.0012 |
| 77 | EMPTY | EMPTY | EMPTY | AC010178.40 | IG51 | (PostTx_En) | (PostTx_Un) | FC = 2.03 | q.value = 0.0234 |