About CORAL
An algorithm has been developed to order fingerprinted clones within contigs. Current version of the CORAL algorithm is written in Java and embedded in iCE (Internet Contig Explorer v. 3.5 by Chris Fjell). Stand-along version of CORAL also available.
Main features:
- Variable tolerance
- Band confirmation
- Band accounting, shared bands
- Vanonical clones identification, based on “fat band” technology
- Two stages burying procedure
- Contig extension algorithm based on “best friend” determination
- Contig evaluation functions, based on quantity of unconfirmed bands in the middle clone, pairwise comparison and Sulston score
- Ability to work with an FPC-file through iCE import-export functions
Testing results on different maps
Leishmania
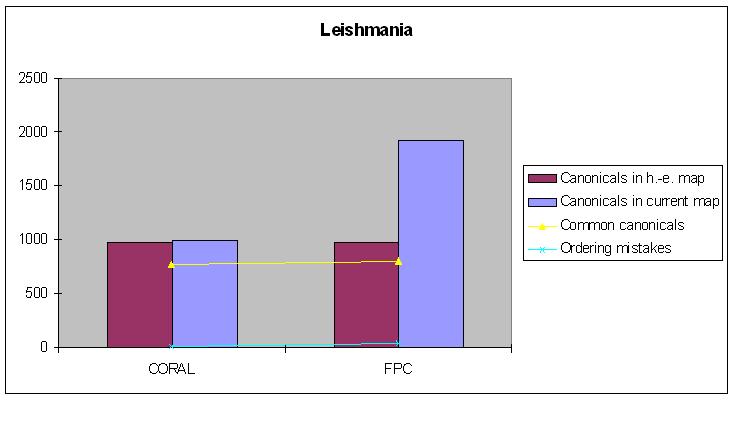
Hand-edited map: 983 canonical clones.
| Canonical clones | Common canonicals | Ordering mistakes | |
|---|---|---|---|
| FPC-map | 1915 | 866 | 46 |
| CORALed map | 994 | 839 | 12 |
JEC21
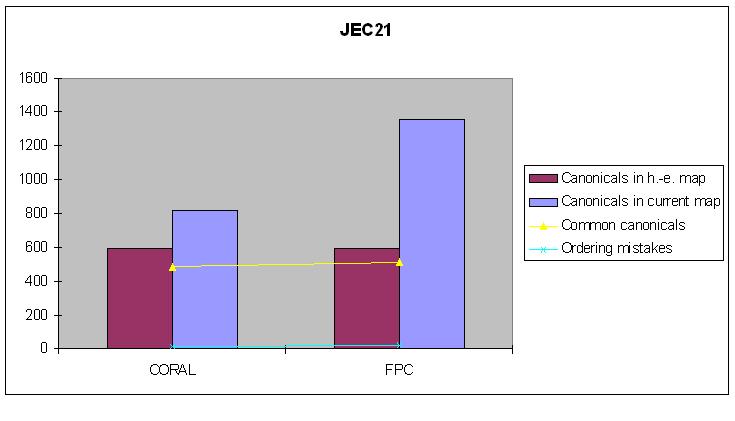
Hand-edited map: 590 canonical clones.
| Canonical clones | Common canonicals | Ordering mistakes | |
|---|---|---|---|
| FPC-map | 1353 | 530 | 23 |
| CORALed map | 818 | 503 | 12 |
Poplar
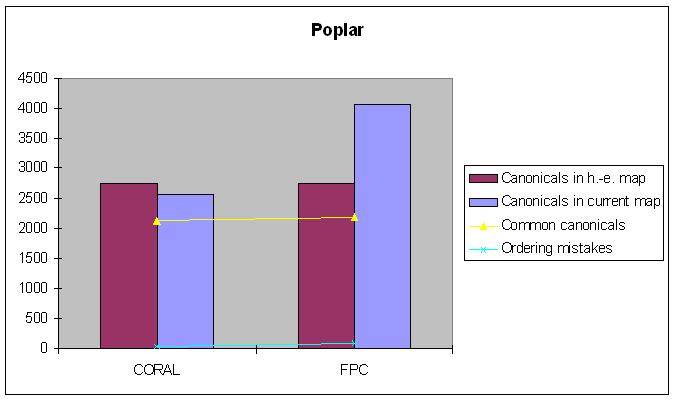
Hand-edited map: 2752 canonical clones.
| Canonical clones | Common canonicals | Ordering mistakes | |
|---|---|---|---|
| FPC-map | 4060 | 2609 | 88 |
| CORALed map | 2555 | 2554 | 29 |
Rhodococcus
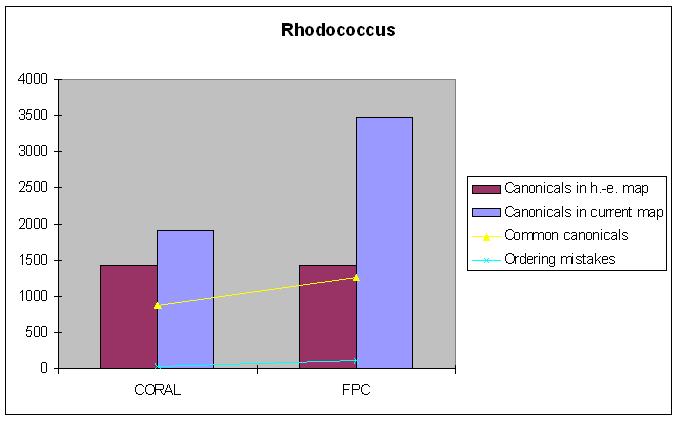
Hand-edited map: 1428 canonical clones.
| Canonical clones | Common canonicals | Ordering mistakes | |
|---|---|---|---|
| FPC-map | 3467 | 1321 | 110 |
| CORALed map | 1913 | 1026 | 26 |