Illustrations¶
Fusion Diagrams¶
These are diagrams produced during the annotate step. These represent the putative fusion events of a single breakpoint pair.
Figure 2. Fusion from transcriptome data. Intronic breakpoints here indicate retained intron sequence and a novel exon is predicted.
Figure 3. Transcriptome breakpoints in the intron result in retained intron and predicts a novel exon.
If the draw_fusions_only flag is set to False then all events will produce a diagram, even anti-sense fusions
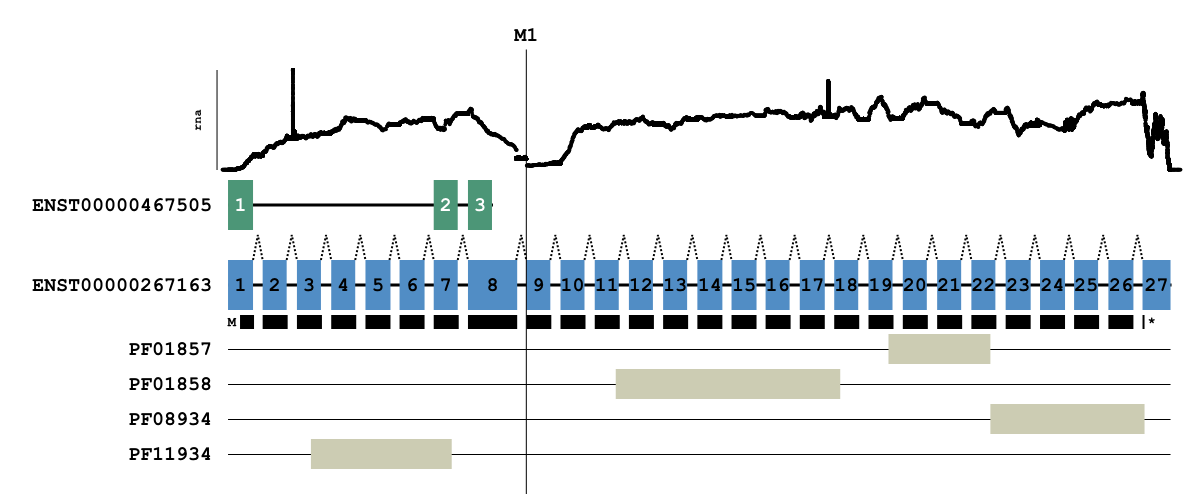
Transcript Overlays¶
MAVIS supports generating diagrams of all transcripts for a given gene. These can be overlaid with markers and bam_file pileup data. This is particularly useful for visualizing splice site mutations.
The above diagram was generated using the overlay command
mavis overlay RB1 \
-o /path/to/output/dir \
--read_depth_plot rna /path/to/bam/file \
--marker M1 48939029 \
--annotations /path/to/mavis/annotations/reference/file