Gene Expression Bioinformatics
Team
Canada's Michael Smith Genome Sciences Centre
BC Cancer
Research Centre
BC Cancer Agency
Vancouver, BC,
Canada
Copyright©2003
|
Version |
Date |
Author |
Comment |
|---|---|---|---|
|
0.1 |
05-May-2003 |
Chris Fjell (cfjell@bcgsc.ca) |
Initial draft. |
|
0.2 |
21-Jul-2003 |
Scott Zuyderduyn (scottz@bcgsc.ca) |
|
|
1.0 |
11-Sept-2003 |
Chris Fjell (cfjell@bcgsc.ca) |
Final draft |
This document describes the operation and maintenance of the DISCOVERY Platform software system. The intended audience consists of end-users and administrators of the DISCOVERY Platform system.
You may see these icons marking text within the documentation:
If you already have a good idea of the purpose and use of the DISCOVERY Platform, you can jump to Client Application Installation (Section 8.1) and then to the How-To chapter (XXX not written yet). However, all users are encouraged to read this document completely – not all features of the DISCOVERY Platform are obvious.
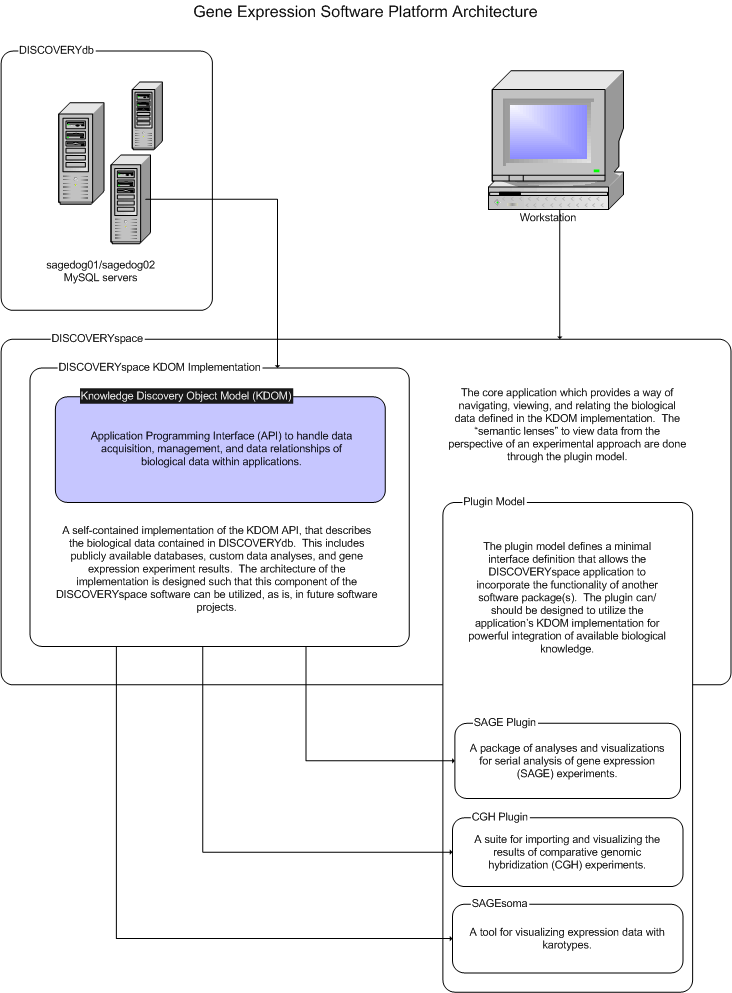
The DISCOVERY Platform is a comprehensive set of software tools to store, visualize, and manipulate genomic data. The Platform consists of several components:
DISCOVERYdb is a database system for acquiring data from public databases, and parsing them to a standard, relation database structure. Experimental data may also be loaded from flat-file formats. The current DISCOVERYdb system uses a MySQL RDBMS and a Java application for acquiring and processing the data.
DISCOVERYspace is the main application for querying and visualizing the data contained in a DISCOVERYdb implementation. Within DISCOVERYspace, data retrieved from DISCOVERYdb are manipulated as datasets.
DISCOVERYspace plugins are optional extensions to the DISCOVERYspace application; these provide experiment- and analysis-specific extensions to the application.
This document is organized around the feature set of the DISCOVERY Platform, not from a perspective of use for a specific purpose. The user is encouraged to read through the How-To chapter (XXX not written yet).
Figure
5.1 DISCOVERY Platform
Minimum
Operating System with Java 1.4.1 or later installed (Windows NT/2000/XP or Linux)
Intel Pentium or AMD Athlon/Duron CPU or equivalent in processing speed
128Mb RAM
Internet connection
100Mb free hard disk space
Keyboard; Mouse
Recommended
2GHz Intel Pentium IV
1Gb RAM
30Gb hard disk
32Mb video card (e.g. GeForce-2 MX)
High speed internet connection
Keyboard; Mouse
One fast machine with lots of memory and disk space, depending on your anticipated usage.
The following terms are used in this document to refer to DISCOVERY Platform features. These include application-specific terms to clarify software terms for the general user and administrator.
A specific selection of data from a single datasource. This is typically the resulting items matching a search against a datasource.
A set of related data from a single source. For example, LocusLink is a datasource, containing a set of data with relationships between the data fields determined by the administrators of LocusLink.
User actions corresponding to using the left mouse button and right mouse button. Users having other pointing devices will be configured differently: left-click is also sometimes called simply select or primary select; while right-click is also called alternate select. The terms, left- and right-click are sometimes used in this document for brevity. Left-click and right-click buttons may also differ with certain operating systems.
The DISCOVERY Platform client application is DISCOVERYspace. The functionality of the main application is extended through additional (optional) plugins.
If you haven't already installed the Java Runtime Environment 1.4.1 or higher, obtain and install it from http://java.sun.com.
Install the application by running the InstallAnywhere application from http://sage.bcgsc.ca/intranet/content/projects/ds/index.mhtml.
The following plugins are available for extended functionality:
This is the analysis plugin for serial analysis of gene expression data. The plugin is currently bundled with the core DISCOVERYspace (8.1.2) application.
This is the analysis plugin for comparative genomic hybridization experiments. (Availability TBA).
This is the plugin for viewing expression data on a karyotype. (Availability TBA).
This is a plugin for viewing natural language processing data. (Availability TBA).
The DISCOVERYdb database server will need to be installed to provide data to the client applications. To install DISCOVERYdb database server perform the following:
Instructions and software are available from MySQL AB (http://www.mysql.com).
Microsoft Windows Specific
For some reason, MySQL wants to lower-case all table names on Windows. Start the server with "-O lower_case_table_names=0" to fix this (ref. http://www.urbansim.org/docs/greenflash/database_information.html).
The maximum packet size for MySQL is ~1M by default. This will not be large enough to slurp in larger tables. The server .cnf file should be modified to include: "--set-variable = max_allowed_packet=4M"
This is done using the bioDatasource software. This command line tool converts a flat-file datasource into a relational database. The tool is run using the syntax below:
CommandLineMain
[OPTIONS] schemaPath dataPathwhere
the schemaPath is the path to a datasource schema file and the
dataPath is the path to the source data file. in some cases
(for example, when using the GENECARDS format, the source data is
distributed as a directory structure). In these cases the
dataPath should point to the root directory of the directory
structure.|
Option |
Description |
|
-help, -? |
Prints help information for command line usage and exits. |
|
-version |
Prints version information and exits. |
|
-v |
Enables verbose output. |
|
-t |
Runs in test mode. In this mode the database is not used, however, the source data will be parsed as normal. This mode is useful for checking configuration and file format issues. |
Table 8.1 bioDatasource parser command line options
This section describes the different components of the DISCOVERY Platform. If you are familiar with the DISCOVERY Platform software and are primarily interested in learning how to do a specific task, you may want to jump to the next section.
On start-up, the main application will be similar to Figure 9.1 after it has started. The main display areas are visible: the Menu Bar (top of the frame), the Project Toolbar (below the Menu Bar), and the Status Bar (bottom of the frame). These are described in detail below:
Figure 9.1 The DISCOVERYspace main frame.
Located at the bottom of the main frame, this bar indicates the status of the connection to the DISCOVERYdb database server on the left-hand side (the box that says "No connection" in Fig. 9.1), the current memory usage (the white box in Fig. 9.1), and buttons to access various desktops (each desktop is a different main window).
This contains menu items for making detailed changes to the application appearance and database connection, as well performing most routine operations in the application.
The Project Menu contains the following menu items:
|
Icon |
Menu Item |
Shortcut |
Description |
|---|---|---|---|
|
|
New |
Ctrl-N |
Starts a new project. |
|
|
Open |
Ctrl-O |
Opens an existing project from disk. |
|
|
Close |
|
Closes the current project. |
|
|
Save |
Ctrl-S |
Saves the current project to disk using the current project filename. If no filename has been given, the user will be prompted for one. |
|
|
Save As... |
|
Saves the current project to disk under a different filename. |
|
|
Import Data > Data Lists... |
|
Displays a dialog window to load data from delimited text files. |
|
|
Import Data > Knowledge XML... |
|
Displays a dialog window to load data in the DISCOVERYspace native XML format. |
|
|
Properties |
|
Displays a dialog window with tabs for altering application preferences (desktop appearance, colours, plugin settings, etc.) |
|
|
Printer Setup... |
Ctrl-Shift-P |
Displays a dialog window to change the default properties for printing. |
|
|
Quit |
Ctrl-Q |
Exits the application. |
Table 9.1 Project Menu Items
The View Menu contains the following menu items:
|
Icon |
Menu Item |
Shortcut |
Description |
|---|---|---|---|
|
|
Toolbars > |
|
This submenu will contain a list of available toolbars, with a checkbox to signify if the toolbar is currently enabled. DISCOVERYspace plugins can contribute items to this sub menu. If no plugins are currently installed that have toolbars, the submenu will display "No additional toolbars available." |
Table 9.2 View Menu Items
The Data Menu contains buttons that, when selected, provide a dialog box to create a dataset from the data item selected. The Data Menu is laid out in a tree. For example, one can create a dataset of Human Refseq entries, based on keyword, by clicking Data > Gene > Genes with Sequence > Refseq > Human Refseq. For a list of available datasources, see Appendix I.
The Tools Menu contains menu items specific to installed plugins. For example, if the SAGE plugin is installed, this menu will contain an option Tools > SAGE > Search For Tag In Libraries. See the documentation for the specific plugin of interest for more information.
The Help Menu contains the following menu items:
|
Icon |
Menu Item |
Shortcut |
Description |
|---|---|---|---|
|
|
Help |
F1 |
Displays on-line help. |
|
|
About |
|
Displays information about the application. |
|
|
Report Bug |
|
Displays a window for the user to submit a report of a defect, including a copy of the application log. |
|
|
Request Feature |
|
Displays a window for the user to submit a feature request, including a copy of the application log. |
|
|
Show application log |
|
Displays the application log. |
Table 9.3 Help Menu Items
The Project Toolbar provides shortcut access to common operations for managing projects. All of these options can also be accessed in the Menu Bar > Project menu.
|
Toolbar |
Name |
Shortcut |
Description |
|---|---|---|---|
|
|
New |
Ctrl-N |
Starts a new project. |
|
|
Open |
Ctrl-O |
Opens an existing project from disk. |
|
|
Save |
Ctrl-S |
Saves the current project to disk using the current project filename. If no filename has been given, the user will be prompted for one. |
|
|
Save As... |
|
Saves the current project to disk under a different filename. |
Table 9.4 Project Toolbar Buttons
DISCOVERYspace allows you to have multiple desktops that you can use to organize your work. At the bottom right of the main application frame, there is a series of icons that you can use to switch desktops (Fig. 9.2). You can see which desktop you're currently viewing by checking the text printed just above these icons (i.e. "DESKTOP 1" in Fig. 9.2). It's possible to increase or decrease the number of available desktops by changing the application settings (see ?????). If more than four desktops are defined, then a button with a double arrow (>>) (Fig. 9.2) will be displayed. When clicked, you will see a list of additional desktops which you can select.
Figure 9.2 The bottom right of the application main frame has components for quick access to multiple desktops.
DISCOVERYspace also allows you to move currently visible windows to different desktops. Windows which can be manipulated in this way have a distinctive look to their top bar (Fig. 9.3). If you right-click on this top bar, you will get a list of available desktops. If you select a different desktop, the window will be moved to the desktop selected. In order to see this window again, you will need to switch your active desktop (see above) to the one the window was moved to.
You're also able to change the title text for these types of windows. The "Rename Window..." option that appears when you right-click the window's top bar (Fig. 9.3) will result in a dialog where you can change the title text (Fig. 9.4). This will allow you to describe the information being displayed in the window in a more personalized way.
Figure 9.3 Right-clicking on the top bar of DISCOVERYspace windows allows you access to options that can move your window to a different desktop or rename the title text.
Figure 9.4 The Rename Window dialog allows you to specify a new title for the selected window.
One the most powerful features of DISCOVERYspace is the ability to do a wide array of keyword searches on available databases. On the Menu Bar, the Data menu will contain an organized list of searchable databases and datatypes (Fig. 9.5). Selecting an item from this menu will result in a search dialog (Fig. 9.6) described below.
Figure 9.5 The Data menu on the application Menu Bar allows you to search available databases.
When you select an item from the Data menu, the Search Dialog will appear (Figs. 9.6-9.7). This dialog allows you to get data based on keywords in searchable fields. The dialog contains a Search Field combo box that you can use to select what information you wish to search on (Fig 9.6). For example, the LocusLink database has accessions, annotations, chromosome number and others that can be searched.
Figure 9.6 The Search Dialog allows you to describe your search. The Search Field combobox contains the searchable fields for the database of interest.
You use the Search Term text field to enter the keyword you wish to search for. In addition, the Case Sensitive and Exact Match checkboxes allow you to specify if you want capitalization to be respected in the search, and if you want the term to exactly match the value of the field, respectively. Often, additional information about the contents of a particular Search Field will be displayed at the bottom of the Search Dialog (i.e. Fig 9.7 shows "Descriptive text for this entry." to describe the "Annotation" Search Field).
Figure 9.7 The Search Dialog allows you to describe your search. The Search Term contains the value you want to search for, and the Case Sensitive and Exact Match checkboxes allow you to define the type of search. If the All Fields checkbox is selected, all fields are searched.
The Data Viewer is a general widget used to display and manage sets of data (Fig. 9.8). The Data Viewer is the most commonly used component of DISCOVERYspace, and knowing how to use it effectively is vital to getting the most out of the application.
The Data Viewer is centred around primary data, the data that populates the Data Table on a row-by-row basis (for example LocusLink Accession and Annotation entries in Figure 9.8). Initially, the Data Viewer display is populated with two columns of primary data, Accession and Annotation).
Figure 9.8 The Data Viewer is one of the most common components used in DISCOVERYspace.
The Data Viewer is organized into five regions: The Data Table, the Top Bar, Menu Bar, Tool Bar, and Status Bar.
In addition to primary data, the Data Viewer also displays linked data, the data related indirectly to the primary data. For example, Refseq data may be displayed on a Data Viewer originally resulting from a search against LocusLink data. Depending on the relationship between the primary and linked data, more than one piece of linked data may associate with the primary data on a single row in the table. For this reason, linked data appears on the data table as drop-down boxes when the number per row is greater than one. The number preceeding the drop-down box is the number of items in the drop-down box (for example, in the first row of the Human Refseq column in Figure 9.9, there are 5 items).
Figure 9.9 A Data Viewer window with primary and linked data.
Figure 9.10 One cell from the data table composed of a drop-down box of linked data.
The data table rows can be sorted by a clicking on the header of a column in the data table. Clicking a second time inverts the order.
Selected rows may be copied to a new Data Viewer display by dragging the rows while holding down either the left or right/middle mouse button (depending on your pointing device configuration). As well, right-clicking on a column will raise a popup menu to perform the copy. Note that the type of the primary data for the new Data Viewer is determined from the column that originated the copy.
The Top Bar shows the title of the current data set. By default, this is the name of the type of data currently being displayed in the Data Viewer followed by a description of the search that generated the list of data if the display is due to a data search. The Top Bar has some useful features described in section 9.1.4 (Desktop and Window Management).
The Menu Bar contains selections for most of the operations available to the user of the Data Viewer.
The Edit Menu contains the following items:
|
Icon |
Menu Item |
Shortcut |
Description |
|---|---|---|---|
|
|
Copy |
Ctrl-C |
Copies the currently selected data rows to the clipboard. |
|
|
Cut |
Ctrl-X |
Removes the currently selected data rows and moves them to the clipboard. |
|
|
Paste |
Ctrl-V |
Copies the data contained in the clipboard to the current Data Table. |
|
|
Paste Special... |
|
(not yet implemented) |
|
|
Delete |
Del |
Deletes the currently selected rows. |
|
|
Select All Rows |
Ctrl-A |
Selects all rows in the Data Table. |
|
|
Deselect All Rows |
|
Deselects all rows in the Data Table. |
|
|
Select By Keyword... |
Ctrl-F |
Selects rows in the Data Table based on a keyword search. |
|
|
Export Data |
|
Exports selected/all to disk. |
Table 9.5 Edit Menu items
The Relationships Menu lists data types that have contextual relationships with the primary data type of the Data Viewer. These items depend on the type of primary data. Selecting an item will create a new column and populate the rows with the appropriate data.
Figure 9.9 The Relationships Menu
Data fields are additional primary data; the fields that are available are determined by the data source.
Figure 9.10 The Data Fields Menu contains additional information that can be displayed for each entry of primary data.
The Tool Bar contains buttons for convenience to perform actions otherwise performed using the Edit Menu. Currently, Delete and Copy buttons are available.
The Status Bar at the bottom of the Data Viewer displays the number of selected rows and the total number of rows.
The datasources available to the user depend on what the DISCOVERY Platform administrator has made available to the DISCOVERYspace client. This is a list of the datasources which are currently supported in a complete deployment of the DISCOVERY Platform. The datasources can be searched against by clicking the corresponding item from the DISCOVERYspace Menu Bar > Data menu (see section 9.1.2.3) – these data fields are listed as Searchable Fields in the following tables. Additional data fields that are not searchable are listed below as Additional Fields.
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Name |
The name of the allelic variant. |
MYASTHENIC SYNDROME, SLOW-CHANNEL CONGENITAL |
|
Synopsis |
A brief synopsis of the allelic variant. |
Engel et al. (1996) described a 30-year-old female patient with ocular and limb weakness, scoliosis, and a family history consistent with autosomal dominant myasthenia gravis (601462) in 3 generations. The mutation leading to pathology in this patient was a heterozygous asn217-to-lys substitution in the AChR-alpha subunit. Engel et al. (1996) evaluated the pathogenicity of the mutation by engineering the mutation into the corresponding cDNA of mouse AChR and coexpressing it with the wildtype cDNA in HEK fibroblasts. Receptor function was evaluated using patch-clamp studies and ACh binding was measured. These studies revealed that the mutations resulted in an apparent increased affinity for ACh and prolonged AChR activation episodes rendering the receptor channel leaky. |
|
Additional Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Mutation |
Notation denoting specific amino acid, etc. changes. |
CHRNA1, SER269ILE |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
10070 |
|
Annotation |
The annotation for the record. |
ABDOMINAL AORTIC ANEURYSM |
|
Alternate Names |
Synonyms for the disease. |
AAA; AORTIC ANEURYSM, ABDOMINAL; ANEURYSM, ABDOMINAL AORTIC ARTERIOMEGALY, INCLUDED; ANEURYSMS, PERIPHERAL, INCLUDED |
|
Features |
Features of the disease. |
Unknown Inheritance |
|
Overall Synopsis |
A verbose text describing details of the disease. |
Tilson and Seashore (1984) reported 50 families in which abdominal aortic aneurysm had occurred in 2 or more first-degree relatives, mainly males. In 29 families, multiple sibs (up to 4) were affected; in 2 families, 3 generations were affected; and in 15 families, persons in 2 generations were affected. Three complex pedigrees were observed: one in which both parents and 3 sons were affected; one in which a man and his paternal uncle were affected; and one in which a man and his father and maternal great-uncle were affected. In the 'one-generation' families, there were 3 with only females affected, including a set of identical twins. (...etc.) |
|
Clinical Synopsis |
Clinical descriptions of the disease. |
vascular; abdominal aortic aneurysm; generalized dilating diathesis; misc; estimated 11.6-fold increase among persons with an affected first-degree; relative; inheritance; autosomal dominant vs. recessive at an autosomal major locus or multifactorial; col3a1 gene (120180.0004) mutations cause about 2% |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
IPR000981 |
|
Annotation |
The annotation for the record. |
Neurhyp_horm |
|
Entry Type |
The type (ie. domain, family, etc.) of the record. |
Family |
|
Protein Classification |
The classification of the protein |
extracellular; Molecular Function:neurohypophyseal hormone activity |
|
Additional Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Number of Matching Proteins |
Number of matching proteins found to correspond to this record. |
86 |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
PF00004 |
|
Annotation |
The annotation for the record. |
ATPase family associated with various cellular activities (AAA) |
|
Description |
Description of the record |
AAA family proteins often perform chaperone-like functions thatassist in the assembly, operation, or disassembly of proteincomplexes [2]. |
|
Additional Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Identifier |
PFAM identifier |
AAA |
|
Family Type |
The type of functional domain. |
Family |
|
Alignment Type |
The source of the alignment math. |
Clustalw |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Name |
The name of the feature. |
CHAIN |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Name |
The name of the organelle. |
chloroplast |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
2 |
|
Annotation |
The annotation for the record. |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
|
Expressed Tissue |
Tissue of gene expression |
Cell lines; adenocarcinoma; colon; corresponding non cancerous liver tissue; hepatocellular carcinoma; liver |
|
Cytoband |
Location of the gene on chromosome |
8p22 |
|
Name |
Name of the gene |
NAT2 |
|
Chromosome |
Chromosome where gene is found |
8 |
The following organisms have Unigene data: Arabidopsis, Human, Mosquito, Mouse.
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
4507652 |
|
Annotation |
The annotation for the record. |
thiopurine S-methyltransferase |
|
Nucleotide Sequence |
The nucleotide sequence of this record. |
CGGCAACCAGCTGTAAGCGAGGCACGG (...etc) |
|
Alphanumeric Accession |
Accession in alphanumeric format |
NM_000367 |
|
Comment |
General comment |
PROVISIONAL REFSEQ: This record has not yet been subject to final NCBI review. The reference sequence was derived from S62904.1. |
|
Chromosome |
Chromosome where gene is found |
6 |
|
Cytoband |
Location of the gene on chromosome |
6p22.3 |
|
Name |
Name of the gene |
TPMT |
|
Protein Sequence |
The protein sequence of this record. |
MDGTRTSLDIEEYSDTEVQKNQVLTLEEWQDKWV (...etc) |
|
Additional Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Nucleotide Sequence Length |
The length of the nucleotide sequence of this record. |
2742 |
|
Gender |
Gender of the organism for this gene sequence |
|
|
Circular Sequence Flag |
Specifies circular that sequence is circular |
false |
|
Addition Date |
Date added to Refseq |
2000-10-31 |
|
Version |
Refseq version |
1 |
|
Sequence Classification |
Classification of the sequence |
Homo sapiens thiopurine S-methyltransferase (TPMT), mRNA. |
|
Protein Sequence Length |
The length of the protein sequence of this record. |
246 |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
4361 |
|
Annotation |
The annotation for the record. |
pleckstrin homology, Sec7 and coiled/coildomains 2, isoform 1 |
|
Nucleotide Sequence |
The nucleotide sequence of this record. |
GGCGGCGGTGGCTCCCGGGGCGTTTGAGCGGGCTCAC (...etc) |
|
Tissue |
Tissue of gene expression |
Lung, small cell carcinoma |
|
Cloning Vector |
Vector used to clone the gene |
pOTB7 |
|
Protein Sequence |
Amino acid sequence of the protein |
MEDGVYEPPDLTPEERMELENIRRRKQELLVEIQRL (...etc) |
|
I.M.A.G.E. ID |
Clone ID in I.M.A.G.E. Consortium data |
3538580 |
|
Additional Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Nucleotide Sequence Length |
The length of the nucleotide sequence of this record. |
1514 |
|
Protein Sequence Length |
The type (ie. domain, family, etc.) of the record. |
400 |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
ENIGMA |
|
Annotation |
The annotation for the record. |
enigma (LIM domain protein) |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
BGLR_ECOLI |
|
Annotation |
The annotation for the record. |
Arabidopsis thaliana |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
1 |
|
Annotation |
The annotation for the record. |
alpha-1-B glycoprotein |
|
Chromosome |
Chromosome where gene is found |
19 |
|
Confirmation Status |
Whether confirmed or not |
true |
|
Function |
Function of the gene product |
Transcription factor |
|
Locus Type |
Type of locus |
gene with protein product, function known or inferred |
|
Phenotype |
Phenotype |
Alzheimer disease, susceptibility to |
|
Product |
Product of the gene |
alpha-1-B glycoprotein |
|
Curation Status |
Curation status |
REVIEWED |
|
Description |
Description |
The protein encoded by this gene is a plasma glycoprotein of unknown function. The protein shows sequence similarity to the variable regions of some immunoglobulin supergene family member proteins. |
|
Name |
Name of the gene |
A1B; A1BG; ABG; GAB |
|
Variant Summary |
Variant Summary |
Transcript variant a includes the alternate exon IA, but not exon IB and encodes a distinct N-terminus. |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
AC3.2 |
|
Annotation |
The annotation for the record. |
contains similarity to Pfam domain: PF00201 (UDP-glucoronosyl and UDP-glucosyl transferases), Score=174.0, E-value=7.7e-49, N=4 |
|
CDNA |
Name of cDNA |
yk822h07.3 |
|
Confirmation Type |
Confirmation type |
EST |
|
Protein |
Protein |
CE05132 |
|
Locus |
Locus |
sri-20 |
|
PCR Product |
PCR product |
mv_AC3.2 |
|
UTR |
UTR |
5_UTR:AC3.4 |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
4362 |
|
Annotation |
The annotation for the record. |
glutathione reductase (NADPH) |
|
Definition |
Definition |
Catalysis of the reaction: 2 glutathione + NADP+ = glutathione disulfide + NADPH + H+. definition_ |
|
Gene Name |
Gene name |
ABF2; ADT1_HUMAN |
|
Synonyms |
Synonyms |
FK506 binding protein; FKBP |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
9606 |
|
Scientific Name |
The complete, canonical name of the taxonomy entry. |
Homo sapiens |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
circadianPathway |
|
Annotation |
The annotation for the record. |
Circadian Rhythms |
|
Description |
Description of the pathway |
Organisms from flies to humans have daily circadian rhythms... |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
ana02010 |
|
Annotation |
The annotation for the record. |
ABC transporters, prokaryotic |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
DPD2_YEAST |
|
Annotation |
The annotation for the record. |
DNA polymerase delta small subunit (EC 2.7.7.7). |
|
Alternate Accession |
Alternate accession |
P46957 |
|
Comments |
Comments |
|
|
Name |
Name of protein |
HUS2; HYS2; J1427; POL31; SDP5; YJR006W; YJR83.7. |
|
Additional Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Keywords |
Keywords |
;Transferase;;DNA-directed DNA polymerase;;DNA replication;;Nuclear protein; |
|
Last Update |
Last Update of database for this entry |
Tue Oct 16 00:00:00 PDT 2001 |
|
Protein Sequence |
Protein sequence |
MDALLTKFNEDRSLQDENLSQPRTR... |
|
Protein Sequence Length |
Length of protein sequence |
487 |
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
DPD2_YEAST |
|
Annotation |
The annotation for the record. |
DNA polymerase delta small subunit (EC 2.7.7.7). |
|
Alternate Accession |
Alternate accession |
P46957 |
|
Comments |
Comments |
|
|
Name |
Name of protein |
HUS2; HYS2; J1427; POL31; SDP5; YJR006W; YJR83.7. |
(This table allows lookup of localization using indices; it contains a list of localization categories.)
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
1 |
|
Annotation |
The annotation for the record. |
cytoplasmic |
(This table allows lookup of localization using indices; it contains a list of localization categories.)
|
Searchable Fields |
||
|---|---|---|
|
Name |
Description |
Example(s) |
|
Accession |
The accession of the record. |
6 |
|
Annotation |
The annotation for the record. |
peroxisomal |