When displaying potentially thousands
of GO terms related with a given set of biological items it is vital
that there be some concept of 'more related' in order to sort the GO
terms by relevance. To do this the the browser scores the set against
the last selected set of items.
The browser scores each term by how it
is associated with the set of
items and how it is related
to them. Thus
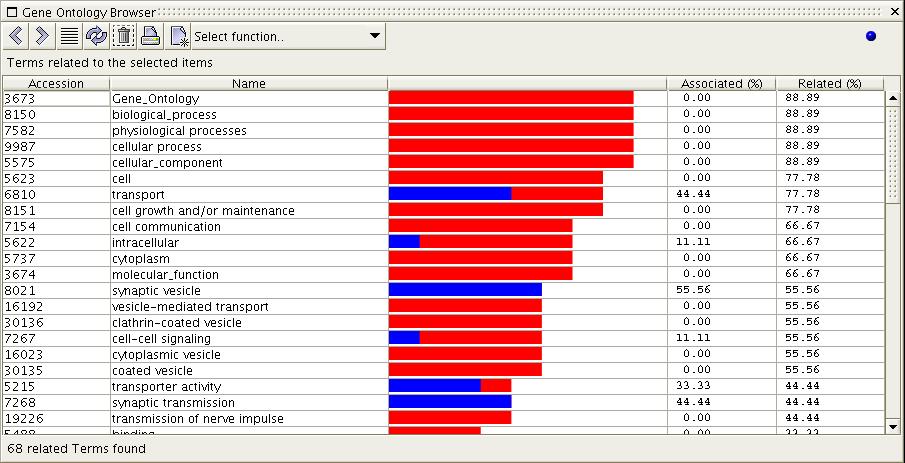
each term is displayed with two scores: Associated(%) and Related(%).
These values are also represented in a bar graph; the blue bar is for
percentage associated and the red bar for percentage related.

Figure
5) In the image above one can
see that the term 'transport' is
related to 77.78% of the last selected items, and is associated with
44.44% of them. The term 'synaptic transmission' has no
relationships other than direct associations. And 'coated vesicle' has
no associations, but is related to over half of the set.
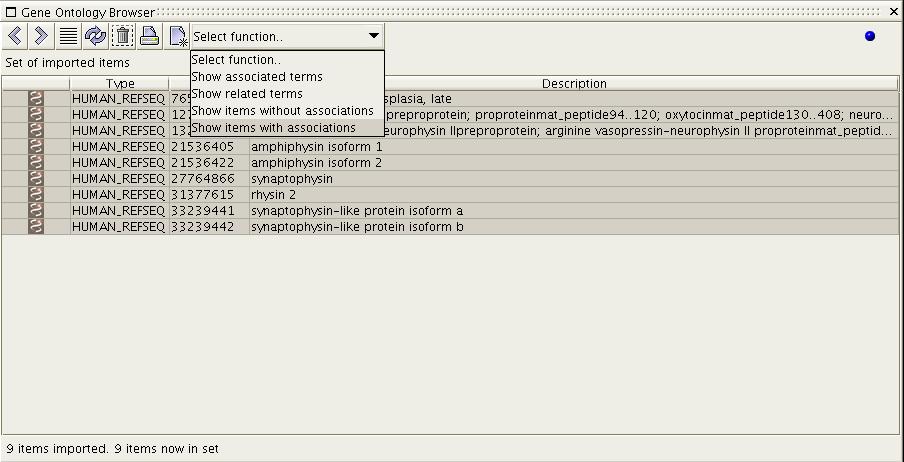
Figure
6a) Previous to the above
screenshot the user has dragged a set
of REFSEQ items onto the browser. She has then isolated 1190 items
using the 'Show items with associations' functions. In this resulting
set she has then selected the first 10 items.
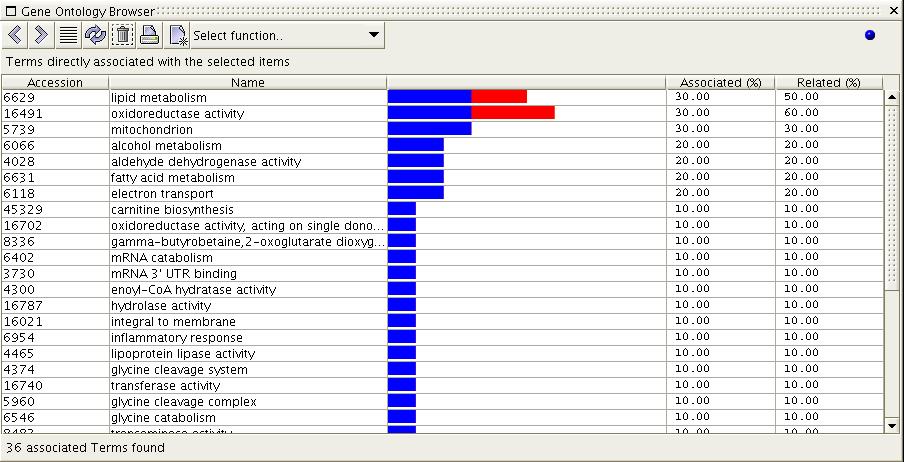
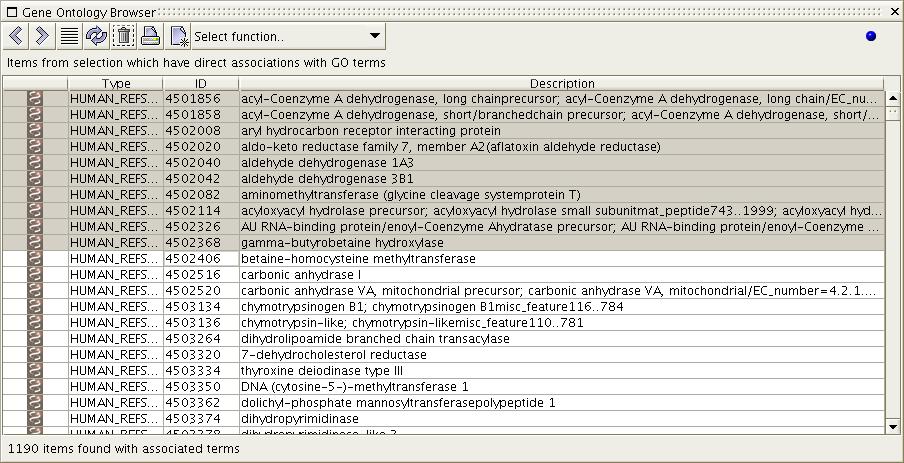
Figure
6b) The image above shows the
result of the 'Show Associated
Terms' function when executed on the selection from figure 6a. All
terms which are directly associated with the selected items are
displayed. Rows are ordered by percentage of association to the
selected items, with the most associated terms first. Notice that the
top two terms have relations to items in addition to the ones that they
are directly associated with.
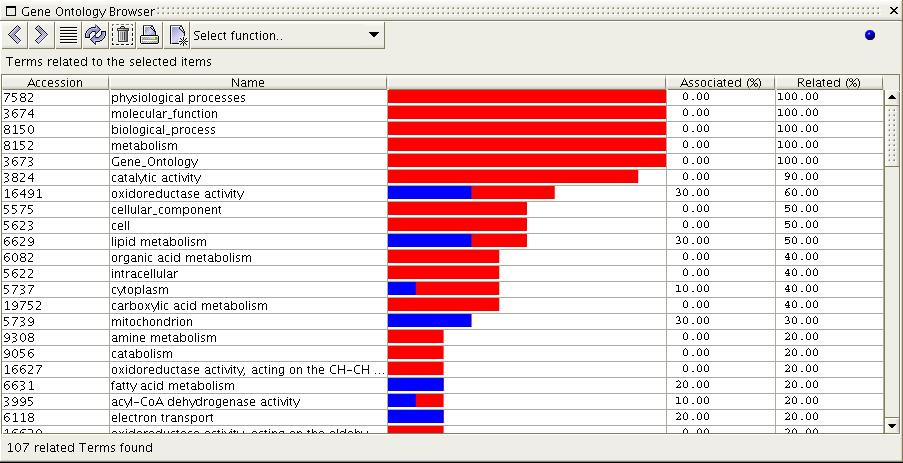
Figure
6c) The image above shows the result of the 'Show Related Terms'
function when executed on the selection from 6a. All terms which are
related to the selected items are displayed. Terms are ordered by
percentage of selected items they are related to, with the most related
terms first. Notice that the root term 'Gene_Ontology' is related to
100% of the items; previously
to figure 6a, all items without associations were removed from the set
using the 'Show items with associations' function.
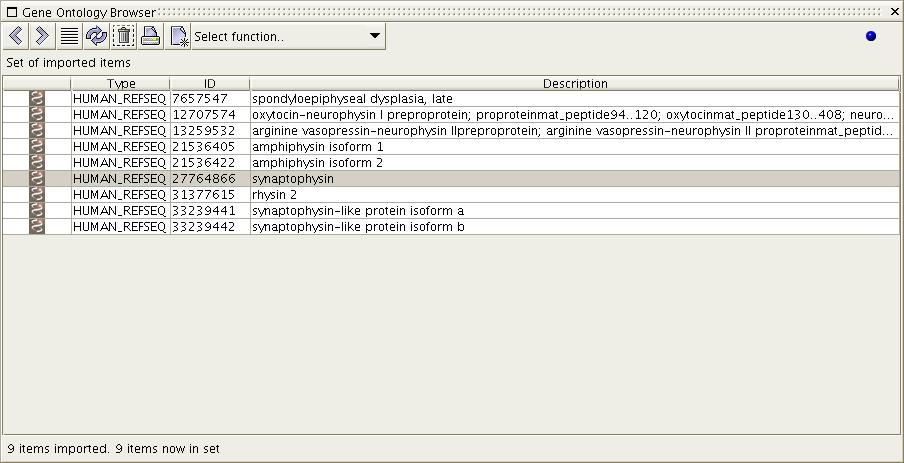
Figure
7a) The figure 6a-c provided
an example case of selecting
multiple items. Notice in this image that only one item has been
selected.
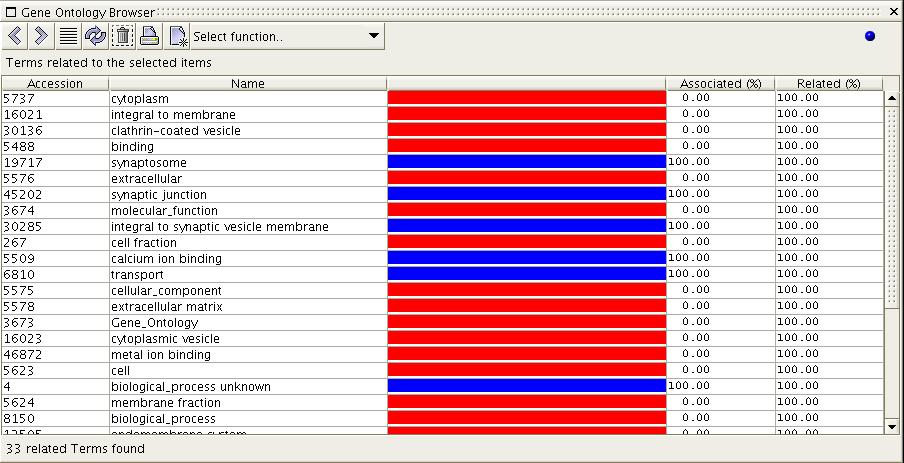
Figure
7b) The image above shows the
result of the 'Show Related Terms'
function when executed on the selection from figure 7a. All terms which
are related to the selected item are displayed. Remember, the scoring
of terms is based upon the last selected set of items; in this case
REFSEQ 27764866 'synaptophysin'. Because there is only one item in the
set, all resulting terms are related to that one term. Thus all terms
are 100% associated/related.