Distributions and lists of all significant differential expression values of the type: SilentIntergenicRegion for comparisons: DLBCL_vs_FL (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Distribution of log2 differential expression values for comparison: DLBCL_vs_FL and data type: SilentIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: SilentIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
Scatter plot of log2 expression values for comparison: DLBCL_vs_FL and data type: SilentIntergenicRegion
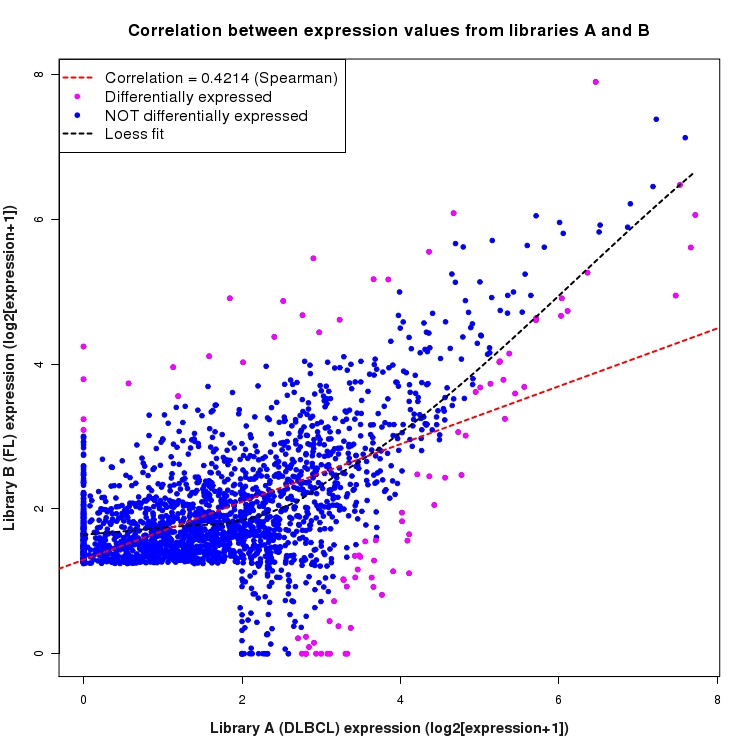
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
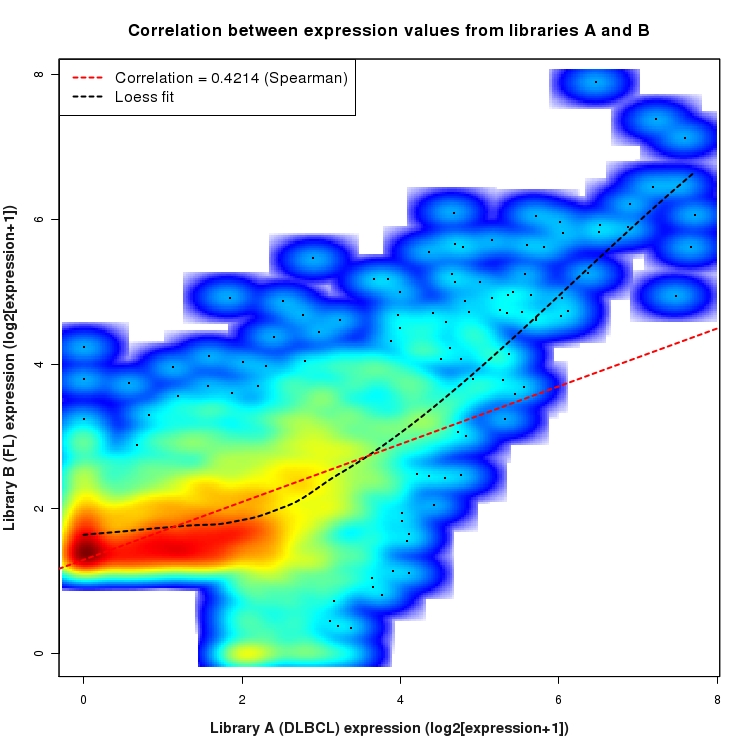
SmoothScatter plot of log2 expression values for comparison: DLBCL_vs_FL and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed SilentIntergenicRegion features
The following table provides a ranked list of all significant differentially expressed SilentIntergenicRegion features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | DLBCL_vs_FL (Gene | Feature | Links | Values) |
| 1 | AC105749.2 U6atac | IG39_SR13 | (DLBCL) | (FL) | FC = -18.95 | q.value = 3.52e-05 |
| 2 | AC016825.12 RP11-539I5.3 | IG49_SR5 | (DLBCL) | (FL) | FC = -13.84 | q.value = 0.00217 |
| 3 | MAP4K3 AC007388.4 | IG34_SR8 | (DLBCL) | (FL) | FC = 10.05 | q.value = 0.00349 |
| 4 | RFTN1 DAZL | IG10_SR2 | (DLBCL) | (FL) | FC = 9.89 | q.value = 0.00349 |
| 5 | INO80B WBP1 | IG11_SR1 | (DLBCL) | (FL) | FC = -9.44 | q.value = 0.0304 |
| 6 | RP11-108A15.2 FXYD8 | IG13_SR16 | (DLBCL) | (FL) | FC = -8.97 | q.value = 0.00282 |
| 7 | AL662795.5 TRIM39 | IG8_SR10 | (DLBCL) | (FL) | FC = 8.64 | q.value = 0.00841 |
| 8 | FAM63B SLTM | IG42_SR1 | (DLBCL) | (FL) | FC = -8.53 | q.value = 0.0377 |
| 9 | ST8SIA4 AC008961.7 | IG24_SR2 | (DLBCL) | (FL) | FC = 8.45 | q.value = 0.0195 |
| 10 | IGHM AL122127.6 | IG19_SR3 | (DLBCL) | (FL) | FC = -8.36 | q.value = 4.14e-06 |
| 11 | C14orf102 | IG1_SR1 | (DLBCL) | (FL) | FC = 8.09 | q.value = 0.0164 |
| 12 | AK2 ADC | IG37_SR1 | (DLBCL) | (FL) | FC = 8.00 | q.value = 0.0195 |
| 13 | AC044802.7 CDH16 | IG21_SR2 | (DLBCL) | (FL) | FC = 7.99 | q.value = 0.000976 |
| 14 | WAC AL161936.15 | IG17_SR1 | (DLBCL) | (FL) | FC = 7.75 | q.value = 0.00265 |
| 15 | SRP_euk_arch AC007384.3 | IG50_SR15 | (DLBCL) | (FL) | FC = 7.65 | q.value = 0.0195 |
| 16 | AK2 ADC | IG37_SR2 | (DLBCL) | (FL) | FC = 7.15 | q.value = 0.0321 |
| 17 | CHST7 SLC9A7 | IG12_SR3 | (DLBCL) | (FL) | FC = -7.10 | q.value = 0.00349 |
| 18 | ADFP RP11-146N23.1 | IG25_SR1 | (DLBCL) | (FL) | FC = 7.00 | q.value = 0.0377 |
| 19 | AC093590.2 AC073043.5 | IG43_SR6 | (DLBCL) | (FL) | FC = 7.00 | q.value = 0.0377 |
| 20 | SOCS1 TNP2 | IG32_SR2 | (DLBCL) | (FL) | FC = 7.00 | q.value = 0.0377 |
| 21 | AMZ1 GNA12 | IG29_SR2 | (DLBCL) | (FL) | FC = 6.81 | q.value = 0.00349 |
| 22 | AL122127.6 AL122127.6 | IG34_SR5 | (DLBCL) | (FL) | FC = 6.77 | q.value = 0.0195 |
| 23 | PHACTR4 RCC1 | IG19_SR1 | (DLBCL) | (FL) | FC = 6.76 | q.value = 0.0377 |
| 24 | AC134980.3 VSIG6 | IG35_SR3 | (DLBCL) | (FL) | FC = 6.74 | q.value = 0.0377 |
| 25 | TNIP1 ANXA6 | IG42_SR5 | (DLBCL) | (FL) | FC = 6.66 | q.value = 0.00481 |
| 26 | AC026150.7 ARHGAP11B | IG50_SR8 | (DLBCL) | (FL) | FC = 6.30 | q.value = 0.0321 |
| 27 | AC011500.7 SUPT5H | IG42_SR4 | (DLBCL) | (FL) | FC = 6.00 | q.value = 0.011 |
| 28 | RP11-666A1.4 NBPF15 | IG6_SR5 | (DLBCL) | (FL) | FC = 5.96 | q.value = 0.0377 |
| 29 | AL122127.6 IGHJ3P | IG20_SR2 | (DLBCL) | (FL) | FC = -5.90 | q.value = 2.18e-08 |
| 30 | TCF12 CGNL1 | IG34_SR7 | (DLBCL) | (FL) | FC = -5.76 | q.value = 0.00481 |
| 31 | AL365361.12 KCNA3 | IG48_SR6 | (DLBCL) | (FL) | FC = 5.76 | q.value = 0.00248 |
| 32 | OR2I1P GABBR1 | IG22_SR1 | (DLBCL) | (FL) | FC = 5.75 | q.value = 6.6e-33 |
| 33 | HFE2 TXNIP | IG30_SR5 | (DLBCL) | (FL) | FC = 5.63 | q.value = 0.0377 |
| 34 | IFI30 AC068499.1 | IG10_SR1 | (DLBCL) | (FL) | FC = 5.50 | q.value = 0.00458 |
| 35 | STT3A CHEK1 | IG28_SR1 | (DLBCL) | (FL) | FC = 5.41 | q.value = 0.0321 |
| 36 | MACROD1 STIP1 | IG9_SR4 | (DLBCL) | (FL) | FC = 5.26 | q.value = 0.0377 |
| 37 | OPRK1 AC022034.7 | IG21_SR7 | (DLBCL) | (FL) | FC = 5.20 | q.value = 0.0138 |
| 38 | AE000660.1 AE000660.1 | IG19_SR4 | (DLBCL) | (FL) | FC = 5.19 | q.value = 0.0224 |
| 39 | STX11 RP1-83M4.2 | IG10_SR2 | (DLBCL) | (FL) | FC = 5.17 | q.value = 0.00213 |
| 40 | TCF12 CGNL1 | IG34_SR8 | (DLBCL) | (FL) | FC = -5.15 | q.value = 0.0321 |
| 41 | ANKRD33B DAP | IG22_SR2 | (DLBCL) | (FL) | FC = -5.11 | q.value = 9.46e-05 |
| 42 | XAB2 PCP2 | IG15_SR3 | (DLBCL) | (FL) | FC = 4.93 | q.value = 0.000388 |
| 43 | HSBP1 MLYCD | IG50_SR3 | (DLBCL) | (FL) | FC = 4.91 | q.value = 0.0419 |
| 44 | FAM65B RP1-245M18.2 | IG19_SR10 | (DLBCL) | (FL) | FC = 4.80 | q.value = 0.0377 |
| 45 | B2M | IG1_SR20 | (DLBCL) | (FL) | FC = 4.76 | q.value = 0.0377 |
| 46 | AE000659.1 AE000659.1 | IG10_SR1 | (DLBCL) | (FL) | FC = 4.56 | q.value = 0.0238 |
| 47 | SLIT2 PACRGL | IG15_SR1 | (DLBCL) | (FL) | FC = 4.45 | q.value = 0.0419 |
| 48 | U1 EOMES | IG47_SR5 | (DLBCL) | (FL) | FC = 4.38 | q.value = 0.00248 |
| 49 | AE000662.1 AE000662.1 | IG22_SR3 | (DLBCL) | (FL) | FC = 4.35 | q.value = 0.0299 |
| 50 | Y_RNA RNF150 | IG50_SR4 | (DLBCL) | (FL) | FC = 4.35 | q.value = 0.0419 |
| 51 | LNPEP AC008865.3 | IG45_SR1 | (DLBCL) | (FL) | FC = 4.21 | q.value = 3.52e-05 |
| 52 | CAMK4 STARD4 | IG7_SR1 | (DLBCL) | (FL) | FC = 4.21 | q.value = 0.0419 |
| 53 | GIMAP1 GIMAP5 | IG30_SR2 | (DLBCL) | (FL) | FC = 4.20 | q.value = 0.0238 |
| 54 | LRAP LNPEP | IG44_SR2 | (DLBCL) | (FL) | FC = 4.14 | q.value = 6.31e-28 |
| 55 | KIAA1618 RNF213 | IG11_SR11 | (DLBCL) | (FL) | FC = -4.03 | q.value = 0.0254 |
| 56 | RP3-415N12.1 TRAF3IP2 | IG5_SR17 | (DLBCL) | (FL) | FC = 4.01 | q.value = 0.0455 |
| 57 | CIITA DEXI | IG28_SR4 | (DLBCL) | (FL) | FC = -3.92 | q.value = 0.00481 |
| 58 | FYCO1 XCR1 | IG44_SR3 | (DLBCL) | (FL) | FC = 3.77 | q.value = 0.0135 |
| 59 | AC105749.2 U6atac | IG39_SR1 | (DLBCL) | (FL) | FC = -3.77 | q.value = 0.00262 |
| 60 | COPS4 PLAC8 | IG19_SR3 | (DLBCL) | (FL) | FC = 3.67 | q.value = 3.21e-05 |
| 61 | BST2 FAM125A | IG38_SR1 | (DLBCL) | (FL) | FC = 3.61 | q.value = 0.000144 |
| 62 | LGALS2 GGA1 | IG42_SR1 | (DLBCL) | (FL) | FC = 3.50 | q.value = 0.00676 |
| 63 | AC020659.5 EHD4 | IG8_SR1 | (DLBCL) | (FL) | FC = 3.33 | q.value = 0.0321 |
| 64 | PTCD2 ZNF366 | IG43_SR13 | (DLBCL) | (FL) | FC = 3.17 | q.value = 0.0108 |
| 65 | RP11-302K17.4 LDB1 | IG27_SR4 | (DLBCL) | (FL) | FC = 3.16 | q.value = 2.09e-20 |
| 66 | NEU3 AP003175.2 | IG49_SR4 | (DLBCL) | (FL) | FC = 2.86 | q.value = 0.00423 |
| 67 | ANKRD33B DAP | IG22_SR3 | (DLBCL) | (FL) | FC = -2.86 | q.value = 0.00248 |
| 68 | ANKRD33B DAP | IG22_SR1 | (DLBCL) | (FL) | FC = -2.76 | q.value = 0.0372 |
| 69 | SREBF2 TNFRSF13C | IG36_SR5 | (DLBCL) | (FL) | FC = -2.71 | q.value = 6.05e-25 |
| 70 | AL122127.6 IGHJ3P | IG20_SR1 | (DLBCL) | (FL) | FC = -2.67 | q.value = 2.73e-06 |
| 71 | RNF115 CD160 | IG41_SR1 | (DLBCL) | (FL) | FC = 2.65 | q.value = 0.0163 |
| 72 | IGHG2 AL928761.2 | IG10_SR6 | (DLBCL) | (FL) | FC = -2.61 | q.value = 0.0224 |
| 73 | SLAMF6 CD84 | IG46_SR4 | (DLBCL) | (FL) | FC = 2.59 | q.value = 0.00023 |
| 74 | TRBV7-2 U66061.1 | IG34_SR1 | (DLBCL) | (FL) | FC = 2.56 | q.value = 0.000454 |
| 75 | KATNAL1 RP11-223E19.1 | IG2_SR11 | (DLBCL) | (FL) | FC = 2.52 | q.value = 0.0377 |
| 76 | FAM120B PSMB1 | IG4_SR16 | (DLBCL) | (FL) | FC = 2.51 | q.value = 0.0362 |
| 77 | CIITA DEXI | IG28_SR2 | (DLBCL) | (FL) | FC = -2.50 | q.value = 0.00349 |
| 78 | P2RX7 P2RX4 | IG24_SR1 | (DLBCL) | (FL) | FC = 2.34 | q.value = 0.0321 |
| 79 | ORAI1 MORN3 | IG31_SR1 | (DLBCL) | (FL) | FC = 2.32 | q.value = 0.0377 |
| 80 | SFXN1 7SK | IG37_SR1 | (DLBCL) | (FL) | FC = 2.32 | q.value = 0.0377 |
| 81 | IGHG4 IGHG2 | IG9_SR6 | (DLBCL) | (FL) | FC = -2.29 | q.value = 0.00262 |
| 82 | P2RY10 RP11-180D15.1 | IG28_SR1 | (DLBCL) | (FL) | FC = 2.18 | q.value = 0.00458 |
| 83 | LRCH3 IQCG | IG30_SR5 | (DLBCL) | (FL) | FC = 2.14 | q.value = 0.0284 |
| 84 | GPR174 RP4-696H22.2 | IG30_SR3 | (DLBCL) | (FL) | FC = 2.14 | q.value = 0.00261 |
| 85 | FGFR3 LETM1 | IG32_SR2 | (DLBCL) | (FL) | FC = 2.11 | q.value = 0.0362 |
| 86 | RP11-302K17.4 LDB1 | IG27_SR5 | (DLBCL) | (FL) | FC = 2.07 | q.value = 6.14e-07 |