Distributions and lists of all significant differential expression values of the type: SilentIntergenicRegion for comparisons: ABC_RG049_vs_GCB_RG069, ABC_vs_GCB (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Differential expression plots for comparison: ABC_vs_GCB and data type: SilentIntergenicRegion
Distribution of log2 differential expression values for comparison: ABC_vs_GCB and data type: SilentIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: SilentIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
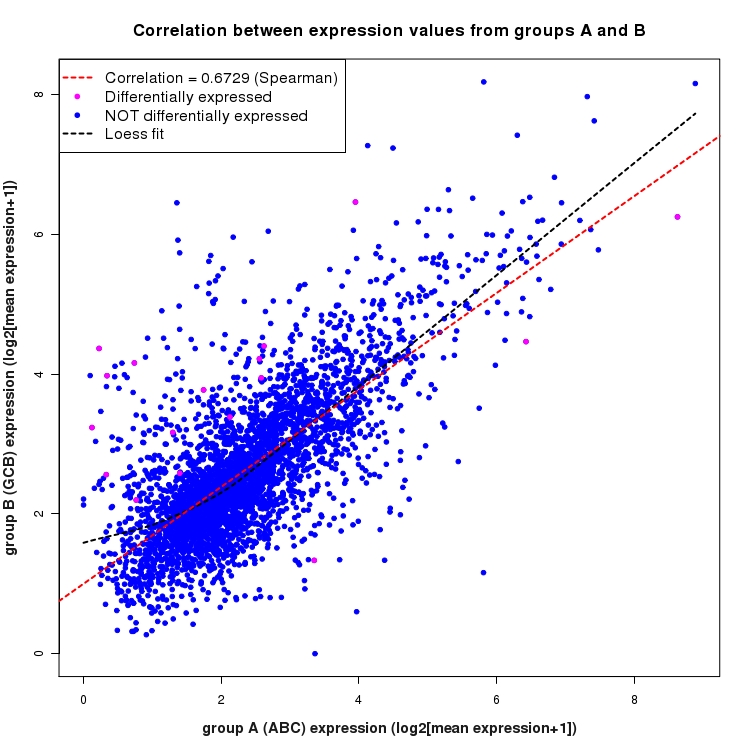
Scatter plot of log2 expression values for comparison: ABC_vs_GCB and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed SilentIntergenicRegion features for groupwise comparisons
The following table provides a ranked list of all significant differentially expressed SilentIntergenicRegion features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | ABC_vs_GCB (Gene | Feature | Links | Values) |
| 1 | RFTN1 RP11-194G10.1 | IG19_SR1 | ABC | GCB | FC = -3.66 | q.value = 0.0195 |
| 2 | 5S_rRNA HCK | IG2_SR2 | ABC | GCB | FC = 4.04 | q.value = 0.0195 |
| 3 | RP11-498B4.2 RP11-539I5.2 | IG50_SR3 | ABC | GCB | FC = -5.73 | q.value = 0.0195 |
| 4 | AC093849.2 HAND2 | IG26_SR14 | ABC | GCB | FC = -4.70 | q.value = 0.0195 |
| 5 | RP11-30O15.1 RP11-44H4.1 | IG19_SR13 | ABC | GCB | FC = -8.66 | q.value = 0.0195 |
| 6 | RP11-109A6.2 RP11-109A6.3 | IG21_SR2 | ABC | GCB | FC = -2.70 | q.value = 0.037 |
| 7 | RP11-30O15.1 RP11-44H4.1 | IG19_SR14 | ABC | GCB | FC = -12.45 | q.value = 0.037 |
| 8 | RP11-613M10.1 FBXO10 | IG12_SR1 | ABC | GCB | FC = -2.28 | q.value = 0.0446 |
| 9 | CCDC65 ARF3 | IG49_SR2 | ABC | GCB | FC = -3.18 | q.value = 0.0446 |
| 10 | CLEC17A AC135052.1 | IG4_SR1 | ABC | GCB | FC = 5.17 | q.value = 0.0449 |
| 11 | PACS1 KLC2 | IG37_SR1 | ABC | GCB | FC = -3.44 | q.value = 0.046 |
| 12 | RP11-30O15.1 RP11-44H4.1 | IG19_SR15 | ABC | GCB | FC = -17.68 | q.value = 0.046 |
| 13 | ICOSLG DNMT3L | IG12_SR4 | ABC | GCB | FC = -10.74 | q.value = 0.046 |
| 14 | C21orf33 AP001055.1 | IG7_SR3 | ABC | GCB | FC = -4.09 | q.value = 0.046 |
| 15 | AL365361.1 AL365361.2 | IG7_SR2 | ABC | GCB | FC = 3.89 | q.value = 0.0473 |
| 16 | EXOC3 AC010442.2 | IG8_SR1 | ABC | GCB | FC = -2.58 | q.value = 0.0473 |
| 17 | AC005003.1 U6 | IG4_SR1 | ABC | GCB | FC = -2.40 | q.value = 0.0473 |
Differential expression plots for comparison: ABC_RG049_vs_GCB_RG069 and data type: SilentIntergenicRegion
Distribution of log2 differential expression values for comparison: ABC_RG049_vs_GCB_RG069 and data type: SilentIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: SilentIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
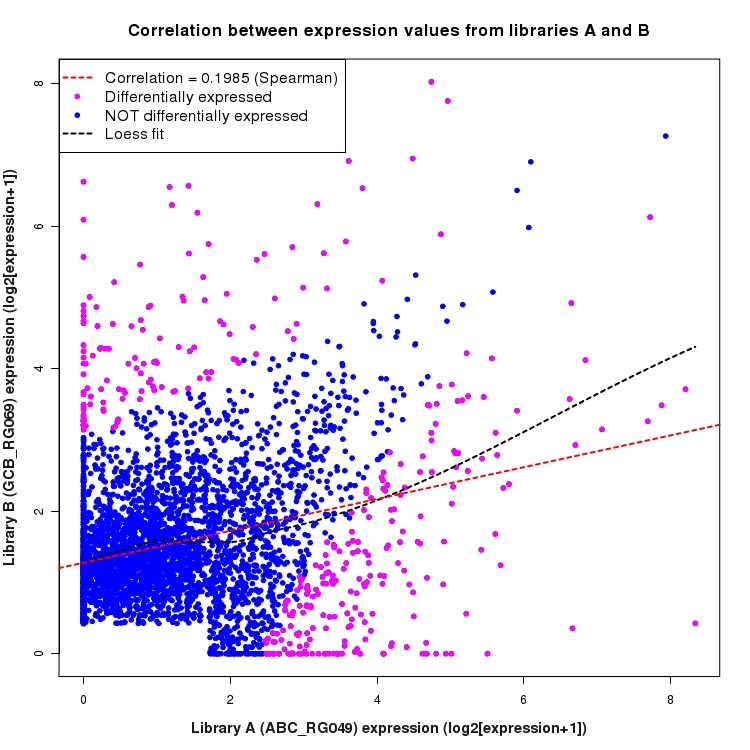
Scatter plot of log2 expression values for comparison: ABC_RG049_vs_GCB_RG069 and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
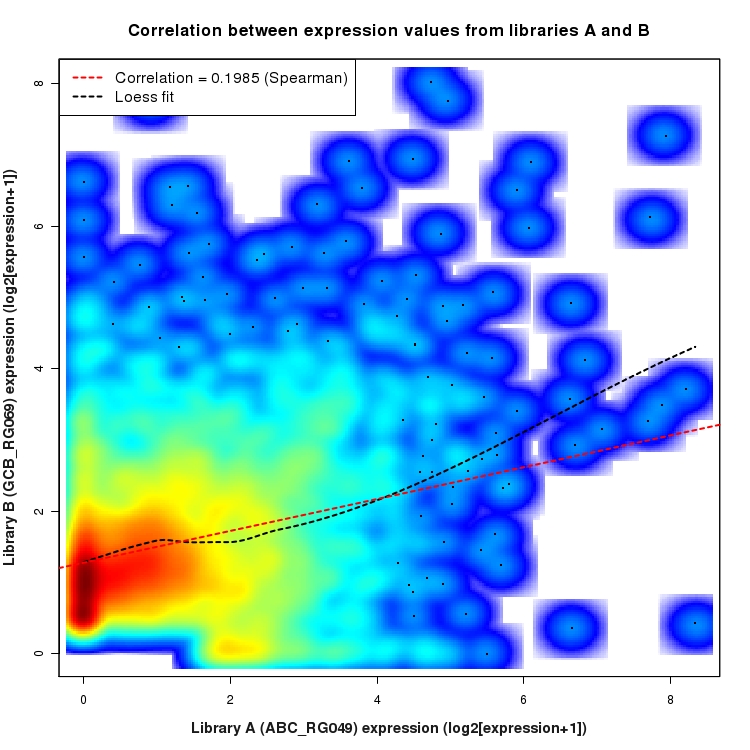
SmoothScatter plot of log2 expression values for comparison: ABC_RG049_vs_GCB_RG069 and data type: SilentIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: SilentIntergenicRegion. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed SilentIntergenicRegion features for pairwise comparisons
The following table provides a ranked list of all significant differentially expressed SilentIntergenicRegion features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | ABC_RG049_vs_GCB_RG069 (Gene | Feature | Links | Values) |
| 1 | CLEC17A AC135052.1 | IG4_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 240.20 | q.value = 2.58e-165 |
| 2 | IGHE IGHG4 | IG21_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -111.85 | q.value = 8.56e-68 |
| 3 | RP13-614K11.1 RP11-374C13.1 | IG17_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -98.81 | q.value = 3.32e-31 |
| 4 | CCR4 GLB1 | IG8_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 79.04 | q.value = 8.18e-49 |
| 5 | RP13-614K11.1 RP11-374C13.1 | IG17_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -68.26 | q.value = 6.88e-21 |
| 6 | ADAR KCNN3 | IG35_SR9 | (ABC_RG049) | (GCB_RG069) | FC = -47.52 | q.value = 3.89e-14 |
| 7 | SNORA67 C6orf105 | IG34_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 45.35 | q.value = 1.46e-21 |
| 8 | RP5-991C6.1 RP11-379B8.1 | IG37_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -41.61 | q.value = 9.33e-28 |
| 9 | ADAR KCNN3 | IG35_SR6 | (ABC_RG049) | (GCB_RG069) | FC = -35.24 | q.value = 1.22e-26 |
| 10 | ADAR KCNN3 | IG35_SR8 | (ABC_RG049) | (GCB_RG069) | FC = -34.19 | q.value = 7.13e-23 |
| 11 | BHLHA15 TECPR1 | IG23_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 32.28 | q.value = 6.28e-15 |
| 12 | BHLHA15 TECPR1 | IG23_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 30.58 | q.value = 2.05e-14 |
| 13 | FCRLB SRP_euk_arch | IG40_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -30.34 | q.value = 4.04e-09 |
| 14 | PRMT8 EFCAB4B | IG32_SR6 | (ABC_RG049) | (GCB_RG069) | FC = -29.67 | q.value = 1.74e-08 |
| 15 | PRR21 OR6B3 | IG14_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -28.04 | q.value = 6.93e-08 |
| 16 | PTK6 SRMS | IG39_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 27.95 | q.value = 6.53e-13 |
| 17 | AL512631.1 ECHDC3 | IG30_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -27.82 | q.value = 8.47e-11 |
| 18 | MTTP DAPP1 | IG33_SR7 | (ABC_RG049) | (GCB_RG069) | FC = -26.77 | q.value = 2.06e-07 |
| 19 | FCRLB SRP_euk_arch | IG40_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -25.86 | q.value = 8.7e-12 |
| 20 | Y_RNA AC097372.1 | IG21_SR10 | (ABC_RG049) | (GCB_RG069) | FC = -25.81 | q.value = 2.77e-08 |
| 21 | AC005682.2 AC005682.4 | IG41_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 25.58 | q.value = 7.01e-12 |
| 22 | MYOM2 CSMD1 | IG16_SR40 | (ABC_RG049) | (GCB_RG069) | FC = -25.49 | q.value = 4.87e-07 |
| 23 | AC079630.3 AC079630.1 | IG24_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 25.33 | q.value = 2.09e-11 |
| 24 | MYNN LRRC34 | IG16_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 25.15 | q.value = 4.42e-16 |
| 25 | AC069113.1 C8orf80 | IG18_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -24.92 | q.value = 1.64e-19 |
| 26 | AC007491.1 AC108084.1 | IG28_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -24.91 | q.value = 4.62e-07 |
| 27 | RASAL2 NCRNA00083 | IG32_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 24.55 | q.value = 2.09e-11 |
| 28 | EIF2AK4 SRP14 | IG4_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 22.88 | q.value = 2.09e-11 |
| 29 | HLA-DRB6 HLA-DRB1 | IG37_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 22.44 | q.value = 1.85e-117 |
| 30 | EIF2AK4 SRP14 | IG4_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 21.69 | q.value = 7.59e-21 |
| 31 | P2RY10 RP11-180D15.1 | IG2_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 21.46 | q.value = 1.19e-81 |
| 32 | GLRX5 TCL6 | IG6_SR9 | (ABC_RG049) | (GCB_RG069) | FC = -21.25 | q.value = 1.18e-06 |
| 33 | TNFSF13B RP11-330C15.1 | IG24_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 20.93 | q.value = 4.47e-93 |
| 34 | Y_RNA AC097372.1 | IG21_SR9 | (ABC_RG049) | (GCB_RG069) | FC = -20.13 | q.value = 1.92e-05 |
| 35 | 7SK ST8SIA4 | IG22_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 19.83 | q.value = 2.26e-09 |
| 36 | MTTP DAPP1 | IG33_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -18.97 | q.value = 4.91e-05 |
| 37 | DEPDC6 COL14A1 | IG48_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -18.78 | q.value = 6.98e-07 |
| 38 | WDR66 BCL7A | IG19_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -18.17 | q.value = 3.96e-12 |
| 39 | MTTP DAPP1 | IG33_SR8 | (ABC_RG049) | (GCB_RG069) | FC = -17.85 | q.value = 0.000133 |
| 40 | BBOX1 CCDC34 | IG7_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 17.02 | q.value = 2.06e-07 |
| 41 | AC079630.3 AC079630.1 | IG24_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 17.00 | q.value = 2.06e-07 |
| 42 | PTK6 SRMS | IG39_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 16.95 | q.value = 6.82e-08 |
| 43 | DOK6 CD226 | IG12_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 16.93 | q.value = 2.06e-07 |
| 44 | MACC1 AC005083.1 | IG16_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -16.83 | q.value = 0.000184 |
| 45 | RP11-292E2.1 RP11-292E2.3 | IG42_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -16.71 | q.value = 3.14e-05 |
| 46 | 5S_rRNA RALGPS2 | IG35_SR9 | (ABC_RG049) | (GCB_RG069) | FC = -16.66 | q.value = 7.8e-05 |
| 47 | FCGR2B RPL31P11 | IG36_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -16.65 | q.value = 3.14e-05 |
| 48 | AL365361.1 AL365361.2 | IG7_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 16.60 | q.value = 6.82e-08 |
| 49 | PRMT8 EFCAB4B | IG32_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -16.53 | q.value = 1.43e-13 |
| 50 | U6 LZTS1 | IG39_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -16.49 | q.value = 0.000184 |
| 51 | ARHGEF1 AC010616.1 | IG24_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -15.84 | q.value = 3.14e-05 |
| 52 | RP11-223E19.2 HMGB1 | IG31_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -15.78 | q.value = 3.77e-07 |
| 53 | AL365361.1 AL365361.2 | IG7_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 15.73 | q.value = 3.13e-09 |
| 54 | ADAR KCNN3 | IG35_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -15.72 | q.value = 3.79e-07 |
| 55 | MDFIC AC068610.2 | IG14_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 15.56 | q.value = 4.64e-16 |
| 56 | SYNE2 ESR2 | IG40_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -15.39 | q.value = 1.38e-05 |
| 57 | IGLV1-44 IGLV7-43 | IG11_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -15.30 | q.value = 3.14e-05 |
| 58 | FAM112A RP5-1030M6.1 | IG18_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 15.25 | q.value = 4.45e-18 |
| 59 | AC099540.2 STT3B | IG42_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 15.22 | q.value = 1.24e-11 |
| 60 | Y_RNA AC097372.1 | IG21_SR8 | (ABC_RG049) | (GCB_RG069) | FC = -15.16 | q.value = 0.000751 |
| 61 | ZNF101 ZNF14 | IG43_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 15.10 | q.value = 1.06e-48 |
| 62 | RP11-498B4.2 RP11-539I5.2 | IG50_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -14.96 | q.value = 5.67e-06 |
| 63 | ZNF101 ZNF14 | IG43_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 13.66 | q.value = 4.13e-36 |
| 64 | AC079630.3 AC079630.1 | IG24_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 13.64 | q.value = 5.96e-06 |
| 65 | RP11-394O9.1 RP11-50K12.1 | IG30_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 13.61 | q.value = 5.96e-06 |
| 66 | DEPDC6 COL14A1 | IG48_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -13.36 | q.value = 2.14e-05 |
| 67 | IL23R AL389925.1 | IG8_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -12.75 | q.value = 0.00308 |
| 68 | Y_RNA AC097372.1 | IG21_SR7 | (ABC_RG049) | (GCB_RG069) | FC = -12.74 | q.value = 0.00283 |
| 69 | hsa-mir-221 hsa-mir-222 | IG11_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 12.73 | q.value = 1.81e-05 |
| 70 | PDK3 PCYT1B | IG9_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -12.68 | q.value = 5.18e-07 |
| 71 | CACNA1D CHDH | IG18_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 12.65 | q.value = 1.81e-05 |
| 72 | CHST7 SLC9A7 | IG26_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -12.61 | q.value = 3.81e-09 |
| 73 | BTBD19 PTCH2 | IG23_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 12.46 | q.value = 5.96e-06 |
| 74 | MTTP DAPP1 | IG33_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -12.46 | q.value = 0.00427 |
| 75 | Y_RNA MAD2L1 | IG8_SR12 | (ABC_RG049) | (GCB_RG069) | FC = 12.37 | q.value = 6.82e-08 |
| 76 | LIPM ANKRD22 | IG40_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 12.25 | q.value = 1e-09 |
| 77 | IL7 RP11-508K19.1 | IG43_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 12.08 | q.value = 1.77e-05 |
| 78 | AC004854.3 MYO1G | IG8_SR6 | (ABC_RG049) | (GCB_RG069) | FC = -12.07 | q.value = 1.2e-06 |
| 79 | RP11-394O9.1 RP11-50K12.1 | IG30_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 11.48 | q.value = 0.000156 |
| 80 | DHRS4L1 LRRC16B | IG9_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -11.46 | q.value = 0.00114 |
| 81 | CDCA7L RAPGEF5 | IG33_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -11.42 | q.value = 0.00427 |
| 82 | MTTP DAPP1 | IG33_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -11.29 | q.value = 0.00672 |
| 83 | CACNA1D CHDH | IG18_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 11.16 | q.value = 0.000156 |
| 84 | PTK6 SRMS | IG39_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 11.13 | q.value = 0.000156 |
| 85 | SLC43A2 SCARF1 | IG25_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -11.11 | q.value = 0.00308 |
| 86 | CLEC2D CLECL1 | IG8_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 11.06 | q.value = 6.82e-08 |
| 87 | U1 U1 | IG34_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -11.05 | q.value = 0.0104 |
| 88 | OS9 AGAP2 | IG12_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -11.00 | q.value = 0.00155 |
| 89 | GALNTL2 DPH3 | IG16_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 10.94 | q.value = 0.000156 |
| 90 | PDK3 PCYT1B | IG9_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -10.93 | q.value = 0.00105 |
| 91 | HMGCL FUCA1 | IG43_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 10.93 | q.value = 0.000156 |
| 92 | RP3-416J7.1 DUSP22 | IG7_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 10.90 | q.value = 5.38e-05 |
| 93 | AC124798.2 KCNJ11 | IG47_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -10.77 | q.value = 0.0166 |
| 94 | XAB2 AC008763.1 | IG31_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 10.66 | q.value = 2.29e-18 |
| 95 | RP11-96L7.1 AL162427.1 | IG5_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 10.63 | q.value = 0.000156 |
| 96 | SEL1L3 C4orf52 | IG39_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -10.63 | q.value = 0.00672 |
| 97 | C4orf29 LARP1B | IG45_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 10.50 | q.value = 5.09e-17 |
| 98 | GBP4 GBP5 | IG9_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 10.47 | q.value = 4.92e-05 |
| 99 | ADAR KCNN3 | IG35_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -10.46 | q.value = 7.81e-05 |
| 100 | SNORD78 VRK2 | IG32_SR20 | (ABC_RG049) | (GCB_RG069) | FC = 10.43 | q.value = 5.38e-05 |
| 101 | U6 GPRIN3 | IG44_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 10.38 | q.value = 1.77e-05 |
| 102 | CCDC65 ARF3 | IG49_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -10.23 | q.value = 0.0165 |
| 103 | RP1-224A6.3 RP1-224A6.2 | IG9_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 10.16 | q.value = 0.00045 |
| 104 | hsa-mir-138-1 C3orf77 | IG14_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -10.10 | q.value = 0.0165 |
| 105 | FAM89B EHBP1L1 | IG15_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 10.09 | q.value = 4.73e-10 |
| 106 | RP11-223E19.2 HMGB1 | IG31_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -9.92 | q.value = 1.2e-06 |
| 107 | CD38 FGFBP1 | IG48_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -9.87 | q.value = 3.31e-25 |
| 108 | AC098614.1 EOMES | IG26_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 9.85 | q.value = 0.00045 |
| 109 | FBXO33 AL390335.1 | IG34_SR10 | (ABC_RG049) | (GCB_RG069) | FC = -9.84 | q.value = 0.0231 |
| 110 | RP11-545P7.1 DTX1 | IG19_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -9.78 | q.value = 2.08e-54 |
| 111 | SNORA67 C6orf105 | IG34_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 9.75 | q.value = 0.00045 |
| 112 | C14orf174 C14orf148 | IG35_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -9.75 | q.value = 0.0039 |
| 113 | PLD4 AHNAK2 | IG5_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -9.73 | q.value = 0.0231 |
| 114 | AC103559.1 U2 | IG26_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -9.63 | q.value = 0.0231 |
| 115 | SCN8A AC068987.1 | IG21_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -9.63 | q.value = 0.0231 |
| 116 | PLXNA3 XX-FW81657B9.1 | IG15_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -9.60 | q.value = 0.0231 |
| 117 | AC092375.1 AC092375.6 | IG29_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -9.59 | q.value = 0.0231 |
| 118 | RP11-30O15.1 RP11-44H4.1 | IG19_SR15 | (ABC_RG049) | (GCB_RG069) | FC = -9.56 | q.value = 0.00672 |
| 119 | AC144836.1 BHLHA9 | IG16_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -9.53 | q.value = 0.00155 |
| 120 | SGK493 AC083949.1 | IG29_SR8 | (ABC_RG049) | (GCB_RG069) | FC = 9.50 | q.value = 0.000364 |
| 121 | AC092933.1 WDR53 | IG12_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 9.42 | q.value = 0.000364 |
| 122 | ATP6V1B2 U6 | IG38_SR6 | (ABC_RG049) | (GCB_RG069) | FC = -9.39 | q.value = 0.00308 |
| 123 | AC079070.1 FBXO15 | IG20_SR6 | (ABC_RG049) | (GCB_RG069) | FC = 9.37 | q.value = 4.92e-05 |
| 124 | PACS1 KLC2 | IG37_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -9.28 | q.value = 0.036 |
| 125 | GALNT8 KCNA6 | IG45_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -9.25 | q.value = 0.036 |
| 126 | MGAT5B AC016168.1 | IG9_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -9.24 | q.value = 0.036 |
| 127 | RP11-44D15.2 BLNK | IG27_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 9.21 | q.value = 9e-07 |
| 128 | AC074290.1 AC107217.1 | IG16_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 9.20 | q.value = 0.00139 |
| 129 | S100Z CRHBP | IG18_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -9.19 | q.value = 0.036 |
| 130 | MORN1 RP4-740C4.2 | IG41_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 9.02 | q.value = 0.000139 |
| 131 | BOD1 CPEB4 | IG9_SR6 | (ABC_RG049) | (GCB_RG069) | FC = -9.01 | q.value = 2.46e-09 |
| 132 | GLRX5 TCL6 | IG6_SR10 | (ABC_RG049) | (GCB_RG069) | FC = -8.99 | q.value = 0.036 |
| 133 | ENTPD4 SLC25A37 | IG27_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -8.94 | q.value = 0.0166 |
| 134 | IGLV1-47 LL22NC03-22A12.1 | IG6_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -8.91 | q.value = 0.00535 |
| 135 | CHST15 OAT | IG37_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -8.88 | q.value = 0.00283 |
| 136 | AEN ISG20 | IG19_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -8.85 | q.value = 2.6e-09 |
| 137 | DOK3 DDX41 | IG5_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -8.84 | q.value = 0.036 |
| 138 | CDCA7L RAPGEF5 | IG33_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -8.79 | q.value = 0.0166 |
| 139 | KIAA1370 ONECUT1 | IG5_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -8.78 | q.value = 0.00155 |
| 140 | NCKAP1L PDE1B | IG6_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -8.73 | q.value = 1.11e-15 |
| 141 | DYNC1H1 AL133223.2 | IG3_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -8.67 | q.value = 0.00104 |
| 142 | STX11 RP1-83M4.1 | IG33_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 8.67 | q.value = 0.00139 |
| 143 | HAVCR2 | IG1_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -8.59 | q.value = 1.34e-06 |
| 144 | PLXNA3 XX-FW81657B9.1 | IG15_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -8.55 | q.value = 0.00283 |
| 145 | 7SK ST8SIA4 | IG22_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 8.52 | q.value = 1.2e-05 |
| 146 | AC018690.1 AFF3 | IG29_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -8.46 | q.value = 0.00155 |
| 147 | RP11-381E24.1 KIAA1524 | IG38_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 8.32 | q.value = 0.000139 |
| 148 | RP11-480N24.2 ZNF318 | IG39_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -8.27 | q.value = 0.00703 |
| 149 | FAM120B PSMB1 | IG34_SR16 | (ABC_RG049) | (GCB_RG069) | FC = 8.27 | q.value = 2.87e-28 |
| 150 | AC010186.2 CLEC2D | IG7_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 8.24 | q.value = 2.06e-07 |
| 151 | RASAL2 NCRNA00083 | IG32_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 8.20 | q.value = 0.0039 |
| 152 | LDB2 SNORA75 | IG5_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 8.19 | q.value = 0.0039 |
| 153 | C1orf146 GLMN | IG45_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 8.15 | q.value = 0.000364 |
| 154 | CTD-2161E19.1 AC008394.1 | IG41_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 8.07 | q.value = 6.8e-06 |
| 155 | PCGF3 AC139887.1 | IG11_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -8.05 | q.value = 0.00028 |
| 156 | AGPS TTC30B | IG30_SR6 | (ABC_RG049) | (GCB_RG069) | FC = 8.03 | q.value = 1.2e-05 |
| 157 | ZNF493 AC010615.2 | IG9_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 7.91 | q.value = 0.0039 |
| 158 | XAB2 AC008763.1 | IG31_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 7.89 | q.value = 5.14e-07 |
| 159 | IL17RA | IG1_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 7.86 | q.value = 0.0039 |
| 160 | GLRX C5orf27 | IG28_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 7.76 | q.value = 0.0039 |
| 161 | AL138756.3 SUSD1 | IG39_SR6 | (ABC_RG049) | (GCB_RG069) | FC = 7.71 | q.value = 0.00281 |
| 162 | AC022826.2 UBE2W | IG20_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 7.69 | q.value = 0.000122 |
| 163 | AC015845.1 MRPS23 | IG32_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -7.64 | q.value = 0.00817 |
| 164 | PPP2R5C | IG1_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 7.64 | q.value = 0.0039 |
| 165 | WDR66 BCL7A | IG19_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -7.56 | q.value = 0.00817 |
| 166 | S1PR4 NCLN | IG33_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 7.55 | q.value = 0.0039 |
| 167 | RP11-359H18.1 ZNF675 | IG35_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 7.52 | q.value = 3.93e-09 |
| 168 | STX11 RP1-83M4.1 | IG33_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 7.46 | q.value = 0.00103 |
| 169 | IGHG2 IGHGP | IG23_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -7.44 | q.value = 0.0179 |
| 170 | C3orf33 SLC33A1 | IG16_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 7.40 | q.value = 0.000216 |
| 171 | AC022511.1 RP11-254B13.1 | IG31_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 7.38 | q.value = 0.0039 |
| 172 | UTS2 TNFRSF9 | IG45_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 7.33 | q.value = 0.000566 |
| 173 | AC008735.1 ZNF784 | IG35_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -7.27 | q.value = 1.89e-09 |
| 174 | KPNA1 | IG1_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 7.23 | q.value = 8.42e-05 |
| 175 | RAI14 TTC23L | IG4_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 7.23 | q.value = 0.011 |
| 176 | UBE2D1 TFAM | IG41_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 7.19 | q.value = 1.66e-13 |
| 177 | USMG5 PDCD11 | IG49_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 7.18 | q.value = 0.000216 |
| 178 | AC022511.1 RP11-254B13.1 | IG31_SR6 | (ABC_RG049) | (GCB_RG069) | FC = 7.18 | q.value = 0.0039 |
| 179 | AC078899.1 ZNF737 | IG48_SR6 | (ABC_RG049) | (GCB_RG069) | FC = 7.15 | q.value = 0.00281 |
| 180 | RP11-231E6.1 BTLA | IG22_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 7.15 | q.value = 0.011 |
| 181 | C9orf46 CD274 | IG2_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 7.08 | q.value = 0.00103 |
| 182 | CR1L CD46 | IG48_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 7.08 | q.value = 0.011 |
| 183 | PLD4 AHNAK2 | IG5_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -7.04 | q.value = 0.0231 |
| 184 | LIPM ANKRD22 | IG40_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 7.04 | q.value = 0.011 |
| 185 | AC106028.2 CHD2 | IG19_SR22 | (ABC_RG049) | (GCB_RG069) | FC = 7.03 | q.value = 8.42e-05 |
| 186 | AC010319.2 FAM125A | IG29_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -7.01 | q.value = 0.0187 |
| 187 | AEN ISG20 | IG19_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -7.00 | q.value = 0.000198 |
| 188 | RAB9A TRAPPC2 | IG13_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -7.00 | q.value = 0.0231 |
| 189 | MED12 NLGN3 | IG31_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -7.00 | q.value = 0.039 |
| 190 | AEN ISG20 | IG19_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -6.96 | q.value = 0.00257 |
| 191 | ZNF574 POU2F2 | IG29_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -6.95 | q.value = 1.41e-38 |
| 192 | LPAR2 GMIP | IG40_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -6.93 | q.value = 0.00159 |
| 193 | RARRES3 HRASLS2 | IG35_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.91 | q.value = 0.011 |
| 194 | RP11-498B4.2 RP11-539I5.2 | IG50_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -6.91 | q.value = 0.0231 |
| 195 | PIK3R5 NTN1 | IG2_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 6.81 | q.value = 0.0015 |
| 196 | RP11-124N19.2 RP11-124N19.1 | IG33_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -6.80 | q.value = 0.036 |
| 197 | S1PR1 RP4-575N6.1 | IG8_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 6.79 | q.value = 0.00703 |
| 198 | AP001636.1 FAM111B | IG4_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -6.78 | q.value = 0.036 |
| 199 | S100A6 S100A5 | IG46_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.76 | q.value = 0.0039 |
| 200 | P2RX7 P2RX4 | IG5_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.71 | q.value = 9.55e-06 |
| 201 | AC004854.3 MYO1G | IG8_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -6.66 | q.value = 0.0179 |
| 202 | GSTZ1 TMED8 | IG33_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -6.66 | q.value = 6.48e-16 |
| 203 | ANKRD60 PPP4R1L | IG21_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 6.62 | q.value = 0.00281 |
| 204 | RP1-164F3.2 TIMM8A | IG34_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -6.57 | q.value = 0.000296 |
| 205 | RP11-659G9.1 RAB30 | IG3_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 6.57 | q.value = 1.54e-28 |
| 206 | ANKRD33B AC106760.1 | IG22_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.53 | q.value = 0.000216 |
| 207 | PHACTR1 AL008729.1 | IG45_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.47 | q.value = 1.59e-08 |
| 208 | TCP11L2 POLR3B | IG33_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.46 | q.value = 3.75e-11 |
| 209 | RP11-62J1.1 RBM28 | IG7_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -6.40 | q.value = 0.0187 |
| 210 | RGS18 RP11-142L4.2 | IG7_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.39 | q.value = 0.011 |
| 211 | CCL3 CCL4 | IG47_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.36 | q.value = 0.011 |
| 212 | GPR174 RP4-696H22.1 | IG4_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 6.35 | q.value = 1.4e-09 |
| 213 | IL17RA | IG1_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.35 | q.value = 0.0285 |
| 214 | RP11-359H18.1 ZNF675 | IG35_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 6.33 | q.value = 0.0285 |
| 215 | AC003090.1 hsa-mir-148a | IG32_SR8 | (ABC_RG049) | (GCB_RG069) | FC = 6.33 | q.value = 2.16e-06 |
| 216 | AL138756.3 SUSD1 | IG39_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 6.33 | q.value = 0.0039 |
| 217 | RP3-416J7.1 DUSP22 | IG7_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 6.28 | q.value = 0.0039 |
| 218 | ANKRD33B AC106760.1 | IG22_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 6.25 | q.value = 0.000148 |
| 219 | RPS27 NUP210L | IG18_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.20 | q.value = 2.96e-05 |
| 220 | C9orf102 NCRNA00092 | IG6_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 6.20 | q.value = 0.011 |
| 221 | ANKRD33B AC106760.1 | IG22_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 6.19 | q.value = 5.97e-05 |
| 222 | C17orf100 SLC13A5 | IG18_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 6.11 | q.value = 0.000294 |
| 223 | NPL AL355999.1 | IG39_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 6.06 | q.value = 0.0285 |
| 224 | CYLC1 RPS6KA6 | IG32_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 6.03 | q.value = 0.0285 |
| 225 | KIAA0391 PSMA6 | IG47_SR10 | (ABC_RG049) | (GCB_RG069) | FC = 6.00 | q.value = 0.0285 |
| 226 | RP11-24F11.2 CCRL2 | IG44_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.83 | q.value = 0.0285 |
| 227 | ANKRD60 PPP4R1L | IG21_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 5.80 | q.value = 0.0285 |
| 228 | RP11-555K12.1 TMEM154 | IG6_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 5.71 | q.value = 2.55e-11 |
| 229 | RBBP8 CABLES1 | IG26_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 5.67 | q.value = 0.0285 |
| 230 | RP5-837D10.1 GBP2 | IG7_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.67 | q.value = 0.0285 |
| 231 | ZNF493 AC010615.2 | IG9_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 5.65 | q.value = 0.0285 |
| 232 | C3orf33 SLC33A1 | IG16_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 5.65 | q.value = 0.000856 |
| 233 | PRMT8 EFCAB4B | IG32_SR5 | (ABC_RG049) | (GCB_RG069) | FC = -5.64 | q.value = 0.0015 |
| 234 | CD47 IFT57 | IG34_SR11 | (ABC_RG049) | (GCB_RG069) | FC = 5.64 | q.value = 7.12e-14 |
| 235 | TRGV5 TRGV4 | IG48_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.62 | q.value = 0.0285 |
| 236 | PIK3CG PRKAR2B | IG24_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -5.53 | q.value = 4.36e-19 |
| 237 | TSG101 UEVLD | IG22_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 5.51 | q.value = 4.91e-08 |
| 238 | AP002358.2 AP002358.1 | IG11_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -5.48 | q.value = 0.0179 |
| 239 | DOCK3 ARMET | IG26_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.45 | q.value = 0.00703 |
| 240 | MIRHG2 C21orf71 | IG40_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.44 | q.value = 0.0285 |
| 241 | U6 AC009283.4 | IG2_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -5.43 | q.value = 0.0179 |
| 242 | snoU13 RP4-575N6.3 | IG6_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 5.42 | q.value = 0.0411 |
| 243 | ELP4 PAX6 | IG27_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.41 | q.value = 0.0411 |
| 244 | AP000872.1 TMEM133 | IG11_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.37 | q.value = 0.00703 |
| 245 | AC108065.1 TIGD2 | IG42_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 5.34 | q.value = 0.00386 |
| 246 | AC106028.2 CHD2 | IG19_SR15 | (ABC_RG049) | (GCB_RG069) | FC = 5.34 | q.value = 0.0285 |
| 247 | PTPLA STAM | IG30_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 5.34 | q.value = 0.0179 |
| 248 | IL15 INPP4B | IG2_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 5.32 | q.value = 0.00386 |
| 249 | hsa-mir-1539 RPL17 | IG45_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 5.30 | q.value = 0.00203 |
| 250 | KPNA1 | IG1_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.25 | q.value = 0.0411 |
| 251 | AC016489.1 POLG2 | IG22_SR15 | (ABC_RG049) | (GCB_RG069) | FC = -5.21 | q.value = 7.23e-05 |
| 252 | AL160008.2 GBP1 | IG5_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 5.21 | q.value = 0.0179 |
| 253 | CD46 MIR29C | IG49_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 5.14 | q.value = 0.00906 |
| 254 | CD28 AC125238.1 | IG13_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.13 | q.value = 0.0179 |
| 255 | SH3BGRL RP11-346E8.1 | IG19_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.12 | q.value = 0.0285 |
| 256 | CD28 AC125238.1 | IG13_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 5.12 | q.value = 0.0211 |
| 257 | FAM65B RP1-245M18.1 | IG2_SR10 | (ABC_RG049) | (GCB_RG069) | FC = 5.12 | q.value = 0.00427 |
| 258 | NCKAP1L PDE1B | IG6_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -5.10 | q.value = 6.93e-07 |
| 259 | DR1 RP4-713B5.1 | IG8_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.07 | q.value = 0.000401 |
| 260 | AC022079.1 CCDC91 | IG21_SR8 | (ABC_RG049) | (GCB_RG069) | FC = 5.06 | q.value = 0.0285 |
| 261 | C4orf34 AC108471.1 | IG39_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 5.01 | q.value = 0.00906 |
| 262 | RP4-552O12.1 PRKACB | IG12_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 5.00 | q.value = 0.0211 |
| 263 | USP38 Y_RNA | IG5_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.95 | q.value = 0.00427 |
| 264 | AC069213.1 AC233280.1 | IG39_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.95 | q.value = 0.0411 |
| 265 | GALNTL2 DPH3 | IG16_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 4.93 | q.value = 0.0285 |
| 266 | POU2AF1 BTG4 | IG32_SR6 | (ABC_RG049) | (GCB_RG069) | FC = -4.93 | q.value = 0.0453 |
| 267 | KLRA1 MAGOHB | IG30_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.92 | q.value = 0.0285 |
| 268 | NOD2 CYLD | IG10_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 4.92 | q.value = 0.01 |
| 269 | ELP4 PAX6 | IG27_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.91 | q.value = 0.00444 |
| 270 | NACA PRIM1 | IG32_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 4.90 | q.value = 2.82e-07 |
| 271 | CST7 C20orf3 | IG39_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.89 | q.value = 0.0285 |
| 272 | RNF34 KDM2B | IG9_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -4.87 | q.value = 0.0166 |
| 273 | PRMT8 EFCAB4B | IG32_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -4.86 | q.value = 0.0231 |
| 274 | RP11-430L16.1 LPP | IG23_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -4.86 | q.value = 0.00179 |
| 275 | CRTAP SUSD5 | IG11_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.86 | q.value = 0.00906 |
| 276 | RPS2P32 Y_RNA | IG3_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.83 | q.value = 0.0179 |
| 277 | DDX20 KCND3 | IG30_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.82 | q.value = 0.0411 |
| 278 | AC022075.1 KLRC4 | IG24_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.82 | q.value = 0.0211 |
| 279 | AL158048.1 hsa-mir-34a | IG17_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 4.81 | q.value = 0.0411 |
| 280 | LSM8 ANKRD7 | IG39_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.77 | q.value = 0.00444 |
| 281 | RP11-555K12.1 TMEM154 | IG6_SR6 | (ABC_RG049) | (GCB_RG069) | FC = 4.76 | q.value = 6.15e-07 |
| 282 | NCKAP1L PDE1B | IG6_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -4.64 | q.value = 3.21e-07 |
| 283 | PLXNB2 FAM116B | IG46_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -4.64 | q.value = 0.0166 |
| 284 | AC007739.1 METTL8 | IG8_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.62 | q.value = 0.000403 |
| 285 | CDKN1B APOLD1 | IG10_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.61 | q.value = 0.0447 |
| 286 | TNFAIP8 5S_rRNA | IG46_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.60 | q.value = 1.32e-06 |
| 287 | AKAP5 ZBTB25 | IG44_SR9 | (ABC_RG049) | (GCB_RG069) | FC = 4.58 | q.value = 1.71e-05 |
| 288 | DCTN6 AC109329.1 | IG39_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.58 | q.value = 0.0218 |
| 289 | IRS1 RHBDD1 | IG21_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 4.53 | q.value = 0.0218 |
| 290 | C3orf33 SLC33A1 | IG16_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.52 | q.value = 0.00906 |
| 291 | AC012100.4 AC012100.1 | IG35_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 4.51 | q.value = 0.00444 |
| 292 | AC078899.1 ZNF737 | IG48_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 4.50 | q.value = 0.0447 |
| 293 | DR1 RP4-713B5.1 | IG8_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.49 | q.value = 0.000932 |
| 294 | TLR1 TLR6 | IG26_SR4 | (ABC_RG049) | (GCB_RG069) | FC = -4.47 | q.value = 0.0231 |
| 295 | AC011471.1 MLLT1 | IG47_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -4.43 | q.value = 0.000145 |
| 296 | NBR1 TMEM106A | IG16_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.42 | q.value = 0.0411 |
| 297 | AP002800.2 MPZL2 | IG19_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.39 | q.value = 0.0411 |
| 298 | AP002360.3 C11orf30 | IG11_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 4.34 | q.value = 0.0447 |
| 299 | SPCS1 NEK4 | IG4_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.28 | q.value = 0.0447 |
| 300 | U6 RP11-458F8.1 | IG9_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -4.27 | q.value = 0.0189 |
| 301 | CLTC PTRH2 | IG11_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.26 | q.value = 0.000401 |
| 302 | IL21R GTF3C1 | IG32_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.24 | q.value = 0.00444 |
| 303 | LSM6 SLC10A7 | IG23_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.19 | q.value = 0.0102 |
| 304 | TXNRD1 CHST11 | IG19_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 4.18 | q.value = 0.0411 |
| 305 | PEX13 AC010733.5 | IG50_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.14 | q.value = 0.0218 |
| 306 | AP000347.4 ZNF70 | IG50_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -4.14 | q.value = 0.0189 |
| 307 | AL356019.1 TTC5 | IG61_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.13 | q.value = 0.0216 |
| 308 | ZNF429 AC092364.1 | IG11_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.12 | q.value = 0.000386 |
| 309 | CLTC PTRH2 | IG11_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.11 | q.value = 0.0411 |
| 310 | C1orf97 RP11-318L16.2 | IG38_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.09 | q.value = 0.000162 |
| 311 | ST6GALNAC4 PIP5KL1 | IG5_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 4.09 | q.value = 0.0411 |
| 312 | RP11-33E24.2 SNHG5 | IG20_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 4.08 | q.value = 0.0411 |
| 313 | ATP13A1 ZNF101 | IG42_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.06 | q.value = 0.0447 |
| 314 | SNX2 SNX24 | IG15_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.05 | q.value = 0.0447 |
| 315 | AL390252.1 PSMA5 | IG24_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 4.05 | q.value = 0.0447 |
| 316 | PRRG4 QSER1 | IG35_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.92 | q.value = 0.0044 |
| 317 | SLC25A3 AC013283.2 | IG16_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -3.91 | q.value = 0.0267 |
| 318 | BST2 AC010319.2 | IG28_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.85 | q.value = 0.01 |
| 319 | NSUN3 RP11-668N23.1 | IG30_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.83 | q.value = 0.0447 |
| 320 | AC006460.1 AC005540.1 | IG4_SR10 | (ABC_RG049) | (GCB_RG069) | FC = 3.81 | q.value = 0.0447 |
| 321 | NIPAL1 TXK | IG35_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 3.79 | q.value = 0.0216 |
| 322 | hsa-mir-607 LCOR | IG36_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 3.78 | q.value = 0.0447 |
| 323 | MACROD1 STIP1 | IG48_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 3.77 | q.value = 0.0429 |
| 324 | THOC7 ATXN7 | IG21_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 3.72 | q.value = 0.0216 |
| 325 | C1orf146 GLMN | IG45_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.69 | q.value = 0.0216 |
| 326 | HN1L MAPK8IP3 | IG13_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 3.65 | q.value = 0.0447 |
| 327 | AEN ISG20 | IG19_SR3 | (ABC_RG049) | (GCB_RG069) | FC = -3.62 | q.value = 0.0403 |
| 328 | Y_RNA TIFA | IG7_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 3.61 | q.value = 0.000338 |
| 329 | ZNF595 | IG1_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 3.60 | q.value = 6.15e-07 |
| 330 | CSNK1G1 KIAA0101 | IG31_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 3.54 | q.value = 0.0092 |
| 331 | CHST7 SLC9A7 | IG26_SR2 | (ABC_RG049) | (GCB_RG069) | FC = -3.51 | q.value = 0.00135 |
| 332 | C4orf29 LARP1B | IG45_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.49 | q.value = 0.0263 |
| 333 | FBXW2 RP11-27I1.3 | IG11_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.41 | q.value = 0.00511 |
| 334 | RP11-98I9.1 USP45 | IG36_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.40 | q.value = 0.0092 |
| 335 | AC007620.1 GNB4 | IG28_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.37 | q.value = 0.0429 |
| 336 | OTUD5 U6 | IG6_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -3.36 | q.value = 0.0221 |
| 337 | RHBDL3 C17orf75 | IG41_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 3.36 | q.value = 0.000245 |
| 338 | Y_RNA MAD2L1 | IG8_SR11 | (ABC_RG049) | (GCB_RG069) | FC = 3.32 | q.value = 0.0205 |
| 339 | RP11-268P4.1 SOX5 | IG34_SR38 | (ABC_RG049) | (GCB_RG069) | FC = -3.32 | q.value = 0.012 |
| 340 | RASSF3 GNS | IG3_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.30 | q.value = 4.45e-14 |
| 341 | AC008074.5 AFTPH | IG36_SR7 | (ABC_RG049) | (GCB_RG069) | FC = 3.19 | q.value = 0.0189 |
| 342 | AC010733.6 PUS10 | IG49_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 3.16 | q.value = 0.00513 |
| 343 | C2orf78 STAMBP | IG40_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 3.12 | q.value = 0.0136 |
| 344 | LSM8 ANKRD7 | IG39_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 3.11 | q.value = 0.000811 |
| 345 | ZNF839 CINP | IG8_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 3.08 | q.value = 3.85e-05 |
| 346 | ZNF839 CINP | IG8_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 3.04 | q.value = 8.42e-05 |
| 347 | RSBN1L TMEM60 | IG27_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.02 | q.value = 0.0347 |
| 348 | RASSF3 GNS | IG3_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 3.01 | q.value = 1.54e-26 |
| 349 | HIST2H2BC AL591493.1 | IG15_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 3.00 | q.value = 0.0136 |
| 350 | ZNF595 | IG1_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 2.97 | q.value = 0.000665 |
| 351 | AC016489.1 POLG2 | IG22_SR13 | (ABC_RG049) | (GCB_RG069) | FC = -2.94 | q.value = 0.0393 |
| 352 | GPR174 RP4-696H22.1 | IG4_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 2.93 | q.value = 0.000362 |
| 353 | GPR174 RP4-696H22.1 | IG4_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 2.92 | q.value = 0.0483 |
| 354 | UBE2V2 AC104989.1 | IG12_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 2.91 | q.value = 0.0285 |
| 355 | ZNF724P AC092329.3 | IG31_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 2.67 | q.value = 4.92e-05 |
| 356 | ZNF595 AC118278.1 | IG2_SR4 | (ABC_RG049) | (GCB_RG069) | FC = 2.55 | q.value = 0.0285 |
| 357 | ZNF595 | IG1_SR6 | (ABC_RG049) | (GCB_RG069) | FC = 2.47 | q.value = 0.00827 |
| 358 | LPIN1 AC096559.2 | IG27_SR3 | (ABC_RG049) | (GCB_RG069) | FC = 2.36 | q.value = 0.00511 |
| 359 | RP11-289I10.2 RP11-289I10.1 | IG10_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 2.34 | q.value = 0.0166 |
| 360 | AC008567.2 ZNF426 | IG39_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 2.31 | q.value = 0.0166 |
| 361 | CFLAR AC007283.2 | IG16_SR1 | (ABC_RG049) | (GCB_RG069) | FC = 2.29 | q.value = 0.0189 |
| 362 | PIK3CG PRKAR2B | IG24_SR1 | (ABC_RG049) | (GCB_RG069) | FC = -2.25 | q.value = 0.0411 |
| 363 | AC019201.2 CYTIP | IG20_SR2 | (ABC_RG049) | (GCB_RG069) | FC = 2.13 | q.value = 0.0236 |
| 364 | MAP1LC3B2 C12orf49 | IG44_SR11 | (ABC_RG049) | (GCB_RG069) | FC = -2.03 | q.value = 0.0128 |
| 365 | RP11-659G9.1 RAB30 | IG3_SR5 | (ABC_RG049) | (GCB_RG069) | FC = 2.00 | q.value = 0.0142 |