Distributions and lists of all significant differential expression values of the type: Intergenic for comparisons: ABC_RG049_vs_GCB_RG069, ABC_vs_GCB (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Differential expression plots for comparison: ABC_vs_GCB and data type: Intergenic
Distribution of log2 differential expression values for comparison: ABC_vs_GCB and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
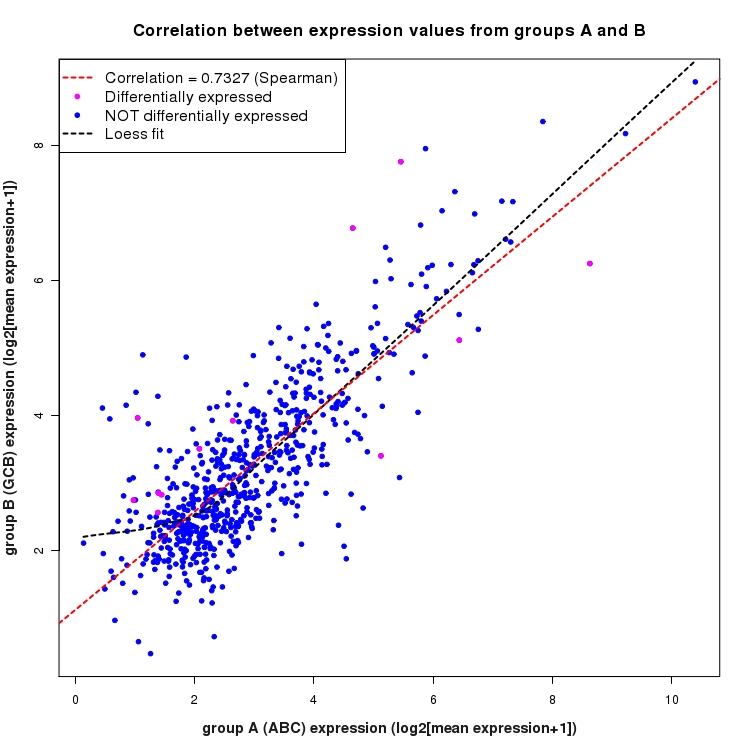
Scatter plot of log2 expression values for comparison: ABC_vs_GCB and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed Intergenic features for groupwise comparisons
The following table provides a ranked list of all significant differentially expressed Intergenic features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | ABC_vs_GCB (Gene | Feature | Links | Values) |
| 1 | AC092143.1 TUBB3 | IG3 | ABC | GCB | FC = -2.76 | q.value = 0.0211 |
| 2 | CLEC17A AC135052.1 | IG4 | ABC | GCB | FC = 5.17 | q.value = 0.0211 |
| 3 | SHISA8 TNFRSF13C | IG28 | ABC | GCB | FC = -4.94 | q.value = 0.0211 |
| 4 | RP11-613M10.1 FBXO10 | IG12 | ABC | GCB | FC = -2.27 | q.value = 0.0211 |
| 5 | PACS1 KLC2 | IG37 | ABC | GCB | FC = -2.70 | q.value = 0.0249 |
| 6 | AL627107.2 SLC25A30 | IG17 | ABC | GCB | FC = 2.49 | q.value = 0.0249 |
| 7 | ICOSLG DNMT3L | IG12 | ABC | GCB | FC = -7.58 | q.value = 0.0249 |
| 8 | AC010323.2 | IG1 | ABC | GCB | FC = -2.61 | q.value = 0.0249 |
| 9 | SSBP3 | IG1 | ABC | GCB | FC = -4.36 | q.value = 0.0406 |
| 10 | NFIX LYL1 | IG11 | ABC | GCB | FC = -3.42 | q.value = 0.0406 |
| 11 | AL365361.1 AL365361.2 | IG7 | ABC | GCB | FC = 3.29 | q.value = 0.0444 |
| 12 | DMPK DMWD | IG31 | ABC | GCB | FC = -2.44 | q.value = 0.0444 |
Differential expression plots for comparison: ABC_RG049_vs_GCB_RG069 and data type: Intergenic
Distribution of log2 differential expression values for comparison: ABC_RG049_vs_GCB_RG069 and data type: Intergenic
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: Intergenic. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
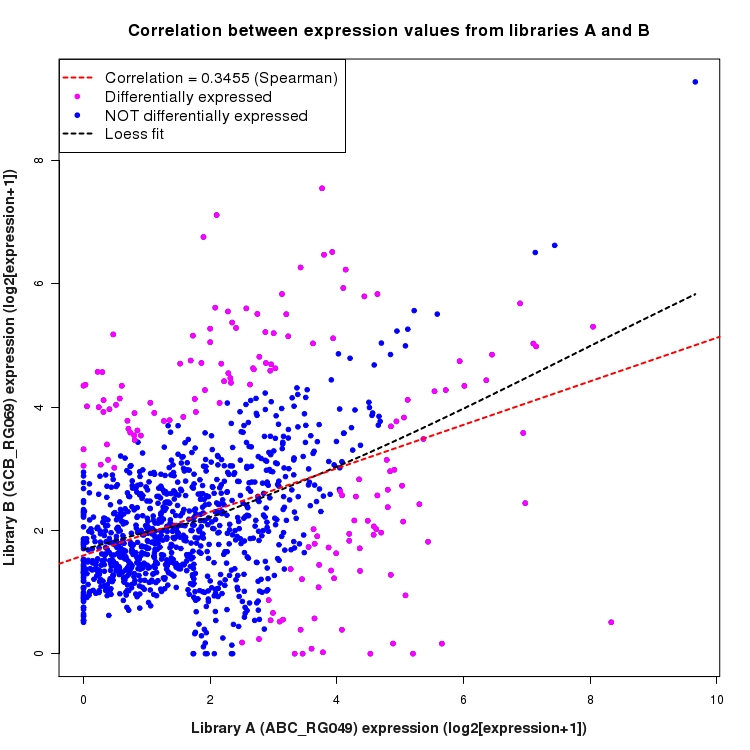
Scatter plot of log2 expression values for comparison: ABC_RG049_vs_GCB_RG069 and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
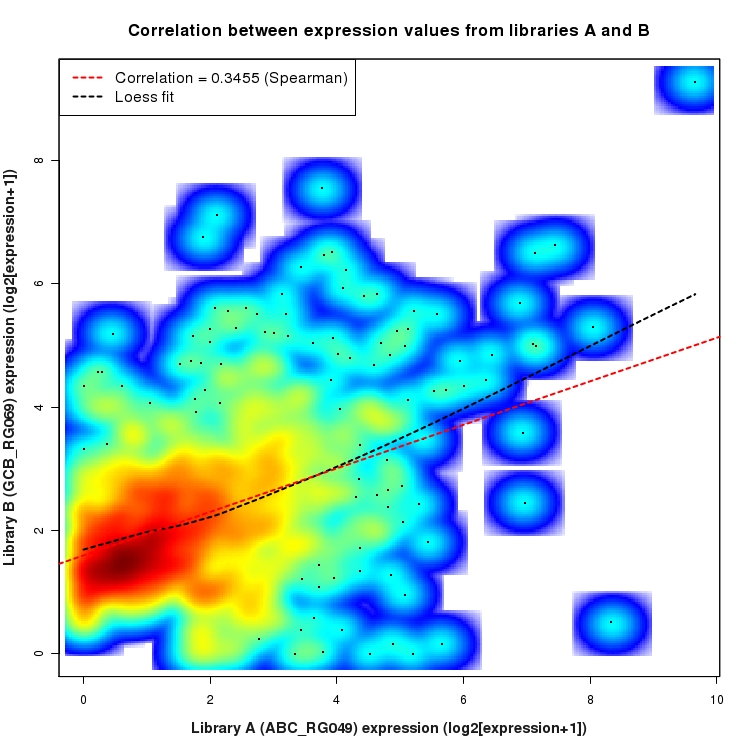
SmoothScatter plot of log2 expression values for comparison: ABC_RG049_vs_GCB_RG069 and data type: Intergenic
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: Intergenic. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed Intergenic features for pairwise comparisons
The following table provides a ranked list of all significant differentially expressed Intergenic features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | ABC_RG049_vs_GCB_RG069 (Gene | Feature | Links | Values) |
| 1 | CLEC17A AC135052.1 | IG4 | (ABC_RG049) | (GCB_RG069) | FC = 226.32 | q.value = 3.68e-145 |
| 2 | AC006299.1 SVIP | IG45 | (ABC_RG049) | (GCB_RG069) | FC = 45.11 | q.value = 9.8e-22 |
| 3 | AC090774.4 USP22 | IG10 | (ABC_RG049) | (GCB_RG069) | FC = 36.81 | q.value = 2.61e-15 |
| 4 | AL110504.2 CCDC85C | IG35 | (ABC_RG049) | (GCB_RG069) | FC = -32.34 | q.value = 7.73e-39 |
| 5 | CCNK AL110504.2 | IG34 | (ABC_RG049) | (GCB_RG069) | FC = -29.17 | q.value = 4.1e-29 |
| 6 | BHLHA15 TECPR1 | IG23 | (ABC_RG049) | (GCB_RG069) | FC = 26.41 | q.value = 3.05e-12 |
| 7 | FCRLB SRP_euk_arch | IG40 | (ABC_RG049) | (GCB_RG069) | FC = -26.16 | q.value = 1.41e-10 |
| 8 | SKIL RP11-469J4.3 | IG26 | (ABC_RG049) | (GCB_RG069) | FC = 23.14 | q.value = 3.8e-45 |
| 9 | SNORA67 C6orf105 | IG34 | (ABC_RG049) | (GCB_RG069) | FC = 23.09 | q.value = 3.06e-09 |
| 10 | FCGR2B RPL31P11 | IG36 | (ABC_RG049) | (GCB_RG069) | FC = -20.39 | q.value = 1.22e-06 |
| 11 | MACC1 AC005083.1 | IG16 | (ABC_RG049) | (GCB_RG069) | FC = -20.36 | q.value = 1.37e-05 |
| 12 | U6 LZTS1 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = -20.16 | q.value = 1.43e-05 |
| 13 | ARHGEF1 AC010616.1 | IG24 | (ABC_RG049) | (GCB_RG069) | FC = -19.32 | q.value = 1.96e-06 |
| 14 | EIF2AK4 SRP14 | IG4 | (ABC_RG049) | (GCB_RG069) | FC = 17.60 | q.value = 1.01e-11 |
| 15 | IL23R AL389925.1 | IG8 | (ABC_RG049) | (GCB_RG069) | FC = -15.55 | q.value = 0.000234 |
| 16 | DHRS4L1 LRRC16B | IG9 | (ABC_RG049) | (GCB_RG069) | FC = -13.90 | q.value = 9.31e-05 |
| 17 | ZNF574 POU2F2 | IG29 | (ABC_RG049) | (GCB_RG069) | FC = -13.77 | q.value = 1.34e-42 |
| 18 | SLC43A2 SCARF1 | IG25 | (ABC_RG049) | (GCB_RG069) | FC = -13.53 | q.value = 0.000234 |
| 19 | CACNA1D CHDH | IG18 | (ABC_RG049) | (GCB_RG069) | FC = 13.52 | q.value = 2.04e-05 |
| 20 | SDF2 SUPT6H | IG27 | (ABC_RG049) | (GCB_RG069) | FC = -13.38 | q.value = 0.000135 |
| 21 | LIPM ANKRD22 | IG40 | (ABC_RG049) | (GCB_RG069) | FC = 12.93 | q.value = 1.08e-05 |
| 22 | TLE4 RP11-79D8.1 | IG26 | (ABC_RG049) | (GCB_RG069) | FC = 12.33 | q.value = 1.09e-13 |
| 23 | RP11-490G8.1 METAP2 | IG36 | (ABC_RG049) | (GCB_RG069) | FC = -12.19 | q.value = 0.000389 |
| 24 | U7 C12orf57 | IG38 | (ABC_RG049) | (GCB_RG069) | FC = 11.90 | q.value = 6.26e-09 |
| 25 | DEPDC6 COL14A1 | IG48 | (ABC_RG049) | (GCB_RG069) | FC = -11.87 | q.value = 9.89e-05 |
| 26 | PLXNA3 XX-FW81657B9.1 | IG15 | (ABC_RG049) | (GCB_RG069) | FC = -11.76 | q.value = 0.000389 |
| 27 | NEK6 PSMB7 | IG9 | (ABC_RG049) | (GCB_RG069) | FC = -11.58 | q.value = 6.93e-11 |
| 28 | RP11-480N24.2 ZNF318 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = -11.50 | q.value = 0.000234 |
| 29 | hsa-mir-221 hsa-mir-222 | IG11 | (ABC_RG049) | (GCB_RG069) | FC = 11.46 | q.value = 0.000135 |
| 30 | RP11-96L7.1 AL162427.1 | IG5 | (ABC_RG049) | (GCB_RG069) | FC = 11.00 | q.value = 0.000351 |
| 31 | EBF1 AC134043.2 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = -10.79 | q.value = 3.89e-08 |
| 32 | OR2I1P UBD | IG7 | (ABC_RG049) | (GCB_RG069) | FC = 10.28 | q.value = 3.95e-35 |
| 33 | RP1-224A6.3 RP1-224A6.2 | IG9 | (ABC_RG049) | (GCB_RG069) | FC = 10.08 | q.value = 0.000912 |
| 34 | MGAT5B AC016168.1 | IG9 | (ABC_RG049) | (GCB_RG069) | FC = -9.96 | q.value = 0.0195 |
| 35 | AEN ISG20 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = -9.67 | q.value = 4.53e-08 |
| 36 | RP11-545P7.1 DTX1 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = -9.66 | q.value = 1.4e-09 |
| 37 | AC018635.1 RP11-62J1.1 | IG6 | (ABC_RG049) | (GCB_RG069) | FC = -9.06 | q.value = 2.55e-05 |
| 38 | WDR66 BCL7A | IG19 | (ABC_RG049) | (GCB_RG069) | FC = -8.56 | q.value = 0.0033 |
| 39 | RP11-124N19.2 RP11-124N19.1 | IG33 | (ABC_RG049) | (GCB_RG069) | FC = -8.49 | q.value = 0.00808 |
| 40 | MORN1 RP4-740C4.2 | IG41 | (ABC_RG049) | (GCB_RG069) | FC = 8.43 | q.value = 0.000368 |
| 41 | ACAA2 SCARNA17 | IG48 | (ABC_RG049) | (GCB_RG069) | FC = -8.36 | q.value = 1.64e-05 |
| 42 | AC008735.1 ZNF784 | IG35 | (ABC_RG049) | (GCB_RG069) | FC = -8.30 | q.value = 1.83e-06 |
| 43 | SERPINA11 SERPINA9 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = -8.28 | q.value = 0.046 |
| 44 | BTBD19 PTCH2 | IG23 | (ABC_RG049) | (GCB_RG069) | FC = 8.22 | q.value = 0.00218 |
| 45 | RMND5B NHP2 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = -8.17 | q.value = 5.34e-09 |
| 46 | SLC25A37 AC051642.2 | IG28 | (ABC_RG049) | (GCB_RG069) | FC = -8.14 | q.value = 4.64e-08 |
| 47 | C12orf52 IQCD | IG23 | (ABC_RG049) | (GCB_RG069) | FC = -8.14 | q.value = 0.0117 |
| 48 | RP11-44D15.2 BLNK | IG27 | (ABC_RG049) | (GCB_RG069) | FC = 8.13 | q.value = 7.54e-06 |
| 49 | AC010319.2 FAM125A | IG29 | (ABC_RG049) | (GCB_RG069) | FC = -8.08 | q.value = 0.00132 |
| 50 | LY9 CD244 | IG4 | (ABC_RG049) | (GCB_RG069) | FC = -7.67 | q.value = 0.0194 |
| 51 | AC009318.1 ERGIC2 | IG27 | (ABC_RG049) | (GCB_RG069) | FC = 7.48 | q.value = 1.56e-08 |
| 52 | SNORD22 SNORD31 | IG16 | (ABC_RG049) | (GCB_RG069) | FC = 7.35 | q.value = 1.4e-09 |
| 53 | GNB2 GIGYF1 | IG24 | (ABC_RG049) | (GCB_RG069) | FC = -7.34 | q.value = 2.96e-07 |
| 54 | AL133351.3 AL133351.1 | IG35 | (ABC_RG049) | (GCB_RG069) | FC = -7.23 | q.value = 9.31e-05 |
| 55 | AP002358.2 AP002358.1 | IG11 | (ABC_RG049) | (GCB_RG069) | FC = -7.21 | q.value = 0.0194 |
| 56 | SHISA8 TNFRSF13C | IG28 | (ABC_RG049) | (GCB_RG069) | FC = -7.15 | q.value = 3.38e-13 |
| 57 | RP11-764K9.4 RP11-764K9.1 | IG23 | (ABC_RG049) | (GCB_RG069) | FC = -6.94 | q.value = 0.046 |
| 58 | MICALL1 C22orf23 | IG13 | (ABC_RG049) | (GCB_RG069) | FC = -6.91 | q.value = 0.0033 |
| 59 | AC010733.8 AC010733.6 | IG48 | (ABC_RG049) | (GCB_RG069) | FC = -6.82 | q.value = 0.0194 |
| 60 | TUBGCP4 TP53BP1 | IG26 | (ABC_RG049) | (GCB_RG069) | FC = -6.81 | q.value = 1.01e-07 |
| 61 | PLD4 AHNAK2 | IG5 | (ABC_RG049) | (GCB_RG069) | FC = -6.76 | q.value = 0.0284 |
| 62 | AL607122.1 FOXD2 | IG24 | (ABC_RG049) | (GCB_RG069) | FC = -6.69 | q.value = 0.0288 |
| 63 | DTX4 MPEG1 | IG8 | (ABC_RG049) | (GCB_RG069) | FC = 6.69 | q.value = 8.02e-61 |
| 64 | IL17RB ACTR8 | IG20 | (ABC_RG049) | (GCB_RG069) | FC = 6.66 | q.value = 1.58e-06 |
| 65 | MYNN LRRC34 | IG16 | (ABC_RG049) | (GCB_RG069) | FC = 6.65 | q.value = 0.000124 |
| 66 | NCKAP1L PDE1B | IG6 | (ABC_RG049) | (GCB_RG069) | FC = -6.50 | q.value = 1.69e-09 |
| 67 | MAP4K4 AC005035.1 | IG2 | (ABC_RG049) | (GCB_RG069) | FC = -6.38 | q.value = 1.58e-14 |
| 68 | AC107375.2 AC107375.1 | IG4 | (ABC_RG049) | (GCB_RG069) | FC = -6.30 | q.value = 0.046 |
| 69 | XAB2 AC008763.1 | IG31 | (ABC_RG049) | (GCB_RG069) | FC = 6.29 | q.value = 2.49e-05 |
| 70 | TMEM49 hsa-mir-21 | IG12 | (ABC_RG049) | (GCB_RG069) | FC = 6.28 | q.value = 7.55e-06 |
| 71 | AC022826.2 UBE2W | IG20 | (ABC_RG049) | (GCB_RG069) | FC = 6.21 | q.value = 0.00151 |
| 72 | ZNF707 AC105219.3 | IG40 | (ABC_RG049) | (GCB_RG069) | FC = -6.20 | q.value = 0.0288 |
| 73 | AL627107.2 SLC25A30 | IG17 | (ABC_RG049) | (GCB_RG069) | FC = 6.07 | q.value = 3.27e-06 |
| 74 | C4orf34 AC108471.1 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = 6.05 | q.value = 0.012 |
| 75 | GSTZ1 TMED8 | IG33 | (ABC_RG049) | (GCB_RG069) | FC = -6.02 | q.value = 1.4e-14 |
| 76 | RP11-381E24.1 KIAA1524 | IG38 | (ABC_RG049) | (GCB_RG069) | FC = 5.96 | q.value = 0.012 |
| 77 | ANKRD33B AC106760.1 | IG22 | (ABC_RG049) | (GCB_RG069) | FC = 5.90 | q.value = 0.000948 |
| 78 | ANKRD60 PPP4R1L | IG21 | (ABC_RG049) | (GCB_RG069) | FC = 5.79 | q.value = 0.0156 |
| 79 | MED12 NLGN3 | IG31 | (ABC_RG049) | (GCB_RG069) | FC = -5.78 | q.value = 0.046 |
| 80 | AC051642.2 AC051642.1 | IG29 | (ABC_RG049) | (GCB_RG069) | FC = -5.77 | q.value = 0.00039 |
| 81 | PHACTR1 AL008729.1 | IG45 | (ABC_RG049) | (GCB_RG069) | FC = 5.73 | q.value = 2.1e-05 |
| 82 | CCDC65 ARF3 | IG49 | (ABC_RG049) | (GCB_RG069) | FC = -5.68 | q.value = 0.00808 |
| 83 | CTD-2324A24.1 | IG1 | (ABC_RG049) | (GCB_RG069) | FC = -5.38 | q.value = 0.0379 |
| 84 | SNORD31 SNORD30 | IG17 | (ABC_RG049) | (GCB_RG069) | FC = 5.38 | q.value = 2.76e-06 |
| 85 | snoU13 GRIP1 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = 5.32 | q.value = 0.027 |
| 86 | FNDC8 NLE1 | IG17 | (ABC_RG049) | (GCB_RG069) | FC = -5.19 | q.value = 0.00473 |
| 87 | TBCE B3GALNT2 | IG45 | (ABC_RG049) | (GCB_RG069) | FC = 5.15 | q.value = 0.000705 |
| 88 | AP000873.1 CCDC90B | IG9 | (ABC_RG049) | (GCB_RG069) | FC = 5.13 | q.value = 0.00124 |
| 89 | RP11-405L18.3 RP11-613M10.1 | IG11 | (ABC_RG049) | (GCB_RG069) | FC = -5.13 | q.value = 0.00626 |
| 90 | DTX1 RASAL1 | IG20 | (ABC_RG049) | (GCB_RG069) | FC = -5.09 | q.value = 1.64e-05 |
| 91 | RPS8 RP11-269F19.2 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = 5.03 | q.value = 9.62e-05 |
| 92 | KPNA1 | IG1 | (ABC_RG049) | (GCB_RG069) | FC = 5.03 | q.value = 0.027 |
| 93 | MIRHG2 C21orf71 | IG40 | (ABC_RG049) | (GCB_RG069) | FC = 5.00 | q.value = 0.0379 |
| 94 | SAMD4B PAF1 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = -4.95 | q.value = 1.43e-06 |
| 95 | ITGA4 CERKL | IG2 | (ABC_RG049) | (GCB_RG069) | FC = 4.94 | q.value = 1.66e-06 |
| 96 | RP11-179H18.3 TIAL1 | IG35 | (ABC_RG049) | (GCB_RG069) | FC = 4.86 | q.value = 0.00457 |
| 97 | AC135457.3 SPATA24 | IG10 | (ABC_RG049) | (GCB_RG069) | FC = -4.81 | q.value = 0.0249 |
| 98 | SLC25A3 AC013283.2 | IG16 | (ABC_RG049) | (GCB_RG069) | FC = -4.80 | q.value = 0.00209 |
| 99 | WWP1 FAM82B | IG50 | (ABC_RG049) | (GCB_RG069) | FC = 4.76 | q.value = 0.000705 |
| 100 | RP11-408A13.1 ZDHHC21 | IG6 | (ABC_RG049) | (GCB_RG069) | FC = 4.72 | q.value = 0.00766 |
| 101 | AKAP14 RP3-327A19.1 | IG32 | (ABC_RG049) | (GCB_RG069) | FC = -4.60 | q.value = 0.00663 |
| 102 | MLL TTC36 | IG27 | (ABC_RG049) | (GCB_RG069) | FC = -4.58 | q.value = 6e-05 |
| 103 | C11orf31 BTBD18 | IG31 | (ABC_RG049) | (GCB_RG069) | FC = 4.47 | q.value = 9.26e-25 |
| 104 | SNORA40 SSH1 | IG8 | (ABC_RG049) | (GCB_RG069) | FC = -4.45 | q.value = 0.00307 |
| 105 | AP003392.2 CCDC84 | IG41 | (ABC_RG049) | (GCB_RG069) | FC = -4.43 | q.value = 0.0167 |
| 106 | TSG101 UEVLD | IG22 | (ABC_RG049) | (GCB_RG069) | FC = 4.43 | q.value = 0.00264 |
| 107 | AL136370.2 UCHL5 | IG22 | (ABC_RG049) | (GCB_RG069) | FC = 4.42 | q.value = 2.14e-05 |
| 108 | PEX13 AC010733.5 | IG50 | (ABC_RG049) | (GCB_RG069) | FC = 4.33 | q.value = 0.00124 |
| 109 | C6orf89 RP1-90K10.1 | IG41 | (ABC_RG049) | (GCB_RG069) | FC = -4.26 | q.value = 4.28e-09 |
| 110 | AD000671.1 TMEM149 | IG44 | (ABC_RG049) | (GCB_RG069) | FC = 4.21 | q.value = 0.000153 |
| 111 | C14orf167 AL136419.1 | IG5 | (ABC_RG049) | (GCB_RG069) | FC = 4.20 | q.value = 2.27e-22 |
| 112 | VPS4A COG8 | IG36 | (ABC_RG049) | (GCB_RG069) | FC = -4.19 | q.value = 0.00663 |
| 113 | AP000872.1 TMEM133 | IG11 | (ABC_RG049) | (GCB_RG069) | FC = 4.15 | q.value = 0.027 |
| 114 | OTUD5 U6 | IG6 | (ABC_RG049) | (GCB_RG069) | FC = -4.12 | q.value = 0.00167 |
| 115 | RP1-164F3.2 TIMM8A | IG34 | (ABC_RG049) | (GCB_RG069) | FC = -3.87 | q.value = 0.0033 |
| 116 | MAVS RP11-119B16.1 | IG25 | (ABC_RG049) | (GCB_RG069) | FC = -3.80 | q.value = 0.0033 |
| 117 | SNORD29 SNORD28 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = 3.79 | q.value = 5.91e-05 |
| 118 | TMX2 C11orf31 | IG30 | (ABC_RG049) | (GCB_RG069) | FC = 3.79 | q.value = 4.01e-12 |
| 119 | LIMK2 PIK3IP1 | IG2 | (ABC_RG049) | (GCB_RG069) | FC = -3.78 | q.value = 0.0193 |
| 120 | MARK2 RCOR2 | IG42 | (ABC_RG049) | (GCB_RG069) | FC = -3.78 | q.value = 0.000283 |
| 121 | FAM118B SRPR | IG16 | (ABC_RG049) | (GCB_RG069) | FC = 3.73 | q.value = 0.0391 |
| 122 | RAB26 SNORD60 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = 3.69 | q.value = 2.74e-06 |
| 123 | MFN1 AC007620.1 | IG27 | (ABC_RG049) | (GCB_RG069) | FC = 3.68 | q.value = 0.000104 |
| 124 | AC007739.1 METTL8 | IG8 | (ABC_RG049) | (GCB_RG069) | FC = 3.65 | q.value = 0.0112 |
| 125 | SSSCA1 FAM89B | IG14 | (ABC_RG049) | (GCB_RG069) | FC = -3.57 | q.value = 0.00341 |
| 126 | CHST7 SLC9A7 | IG26 | (ABC_RG049) | (GCB_RG069) | FC = -3.56 | q.value = 2.37e-06 |
| 127 | ELP4 PAX6 | IG27 | (ABC_RG049) | (GCB_RG069) | FC = 3.54 | q.value = 0.033 |
| 128 | C4orf43 MARCH1 | IG31 | (ABC_RG049) | (GCB_RG069) | FC = 3.45 | q.value = 0.0275 |
| 129 | CLTC PTRH2 | IG11 | (ABC_RG049) | (GCB_RG069) | FC = 3.37 | q.value = 0.00473 |
| 130 | TCIRG1 CHKA | IG5 | (ABC_RG049) | (GCB_RG069) | FC = -3.34 | q.value = 0.0202 |
| 131 | METTL11A | IG51 | (ABC_RG049) | (GCB_RG069) | FC = -3.32 | q.value = 0.0102 |
| 132 | RP11-659G9.1 RAB30 | IG3 | (ABC_RG049) | (GCB_RG069) | FC = 3.18 | q.value = 2.51e-08 |
| 133 | PPP2R3C KIAA0391 | IG46 | (ABC_RG049) | (GCB_RG069) | FC = 3.12 | q.value = 0.000688 |
| 134 | BET1L RIC8A | IG5 | (ABC_RG049) | (GCB_RG069) | FC = -3.12 | q.value = 0.0273 |
| 135 | BST2 AC010319.2 | IG28 | (ABC_RG049) | (GCB_RG069) | FC = 3.06 | q.value = 0.0494 |
| 136 | SFT2D2 ANKRD36BL1 | IG24 | (ABC_RG049) | (GCB_RG069) | FC = -3.03 | q.value = 0.0195 |
| 137 | RPL36 LONP1 | IG34 | (ABC_RG049) | (GCB_RG069) | FC = 3.03 | q.value = 5.68e-10 |
| 138 | AC010733.6 PUS10 | IG49 | (ABC_RG049) | (GCB_RG069) | FC = 2.87 | q.value = 0.015 |
| 139 | RP11-98I9.1 USP45 | IG36 | (ABC_RG049) | (GCB_RG069) | FC = 2.86 | q.value = 0.0244 |
| 140 | GPX3 TNIP1 | IG8 | (ABC_RG049) | (GCB_RG069) | FC = 2.72 | q.value = 1.72e-05 |
| 141 | CPSF7 SDHAF2 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = 2.67 | q.value = 0.0244 |
| 142 | RP1-187N21.1 NUDT3 | IG41 | (ABC_RG049) | (GCB_RG069) | FC = -2.66 | q.value = 0.0113 |
| 143 | AC092473.2 CXorf38 | IG13 | (ABC_RG049) | (GCB_RG069) | FC = -2.57 | q.value = 0.000782 |
| 144 | COX4NB COX4I1 | IG43 | (ABC_RG049) | (GCB_RG069) | FC = 2.44 | q.value = 0.000191 |
| 145 | RAD9B PPTC7 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = 2.34 | q.value = 0.00646 |
| 146 | RPS25 TRAPPC4 | IG42 | (ABC_RG049) | (GCB_RG069) | FC = 2.31 | q.value = 2.11e-09 |
| 147 | MSI2 U7 | IG28 | (ABC_RG049) | (GCB_RG069) | FC = -2.29 | q.value = 0.00239 |
| 148 | MED28 FAM184B | IG12 | (ABC_RG049) | (GCB_RG069) | FC = 2.29 | q.value = 8.22e-05 |
| 149 | PLCG2 7SK | IG8 | (ABC_RG049) | (GCB_RG069) | FC = -2.26 | q.value = 0.0348 |
| 150 | MCM4 U6 | IG10 | (ABC_RG049) | (GCB_RG069) | FC = 2.25 | q.value = 0.00702 |
| 151 | LSM1 BAG4 | IG39 | (ABC_RG049) | (GCB_RG069) | FC = 2.24 | q.value = 0.0129 |
| 152 | SNORND104 SNORA76 | IG19 | (ABC_RG049) | (GCB_RG069) | FC = 2.00 | q.value = 0.0181 |