Distributions and lists of all significant differential expression values of the type: ActiveIntergenicRegion for comparisons: Mip5FuR_vs_Mip101 (p-value < 0.05 after multiple testing correction and Fold-Change > 1.5)
Differential expression plots for comparison: Mip5FuR_vs_Mip101 and data type: ActiveIntergenicRegion
Distribution of log2 differential expression values for comparison: Mip5FuR_vs_Mip101 and data type: ActiveIntergenicRegion
Distribution of all differential expression values that meet the p-value and fold-change cutoff for the feature type: ActiveIntergenicRegion. The total number of significant DE features, as well as the max and min log2 DE observed are noted in the legend. *If you can not see the figure below, click here
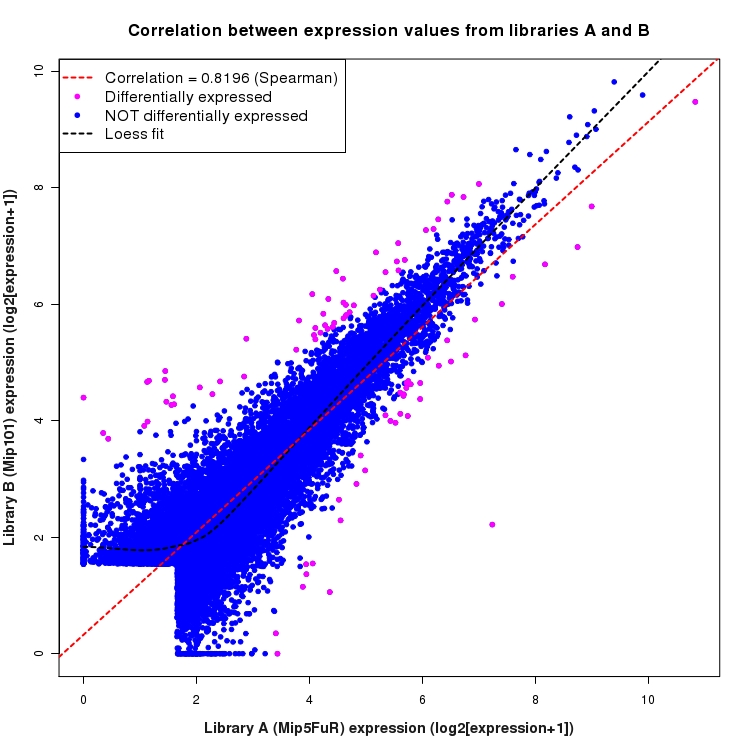
Scatter plot of log2 expression values for comparison: Mip5FuR_vs_Mip101 and data type: ActiveIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: ActiveIntergenicRegion. Features that are differentially expressed (meet the p-value and fold-change cutoff) are indicated in magenta. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
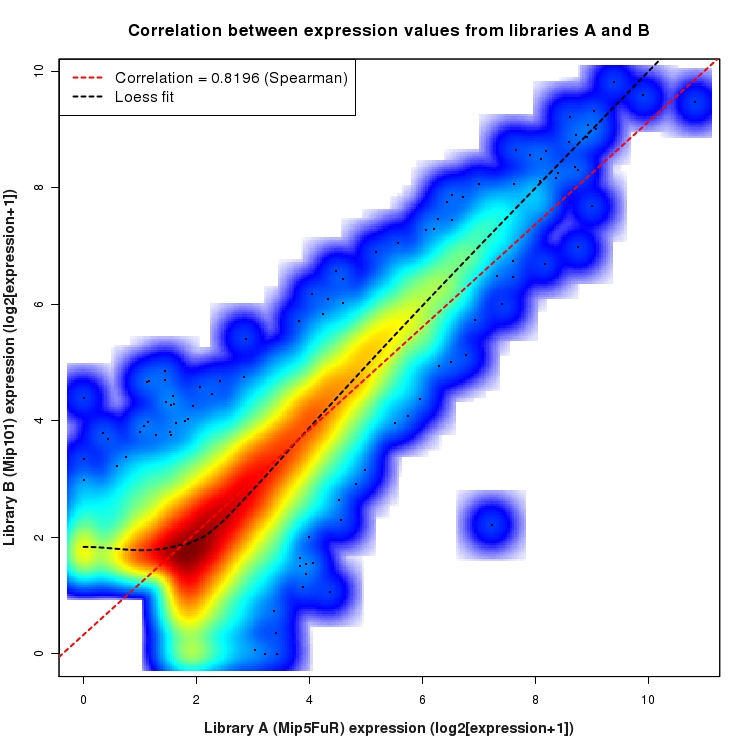
SmoothScatter plot of log2 expression values for comparison: Mip5FuR_vs_Mip101 and data type: ActiveIntergenicRegion
Correlation between expression values for all features that are expressed above background in one or both libraries for the feature type: ActiveIntergenicRegion. A linear model is fit to the data and the correlation by Spearman method is reported. A loess model is also fit to illustrate the trend of the data.
Significant differentially expressed ActiveIntergenicRegion features
The following table provides a ranked list of all significant differentially expressed ActiveIntergenicRegion features. Each column corresponds to a pair-wise comparison of two libraries. Each table cell contains the Gene Name (which links to the ALEXA-Seq gene record), the Feature Name, the name of each library being compared (which links to the feature's coordinates in the UCSC Genome Browser and displays expression data), the fold-change (FC) of the differential expression event, and the multiple testing corrected p-value for the feature. A bold row indicates that the feature is not currently supported by EST or mRNA sequence alignments. For exon junction features the number of exons skipped by the junction is indicated as 'Sn' where n is the number of exons skipped (e.g. S0 means no exons skipped, S1 means one exon skipped, etc.).
| RANK | Mip5FuR_vs_Mip101 (Gene | Feature | Links | Values) |
| 1 | AC034236.5 COMMD10 | IG34_AR3 | (Mip5FuR) | (Mip101) | FC = 32.54 | q.value = 2.89e-51 |
| 2 | MLNR CDADC1 | IG10_AR1 | (Mip5FuR) | (Mip101) | FC = -21.07 | q.value = 8.32e-05 |
| 3 | HHIP AC109811.4 | IG15_AR6 | (Mip5FuR) | (Mip101) | FC = -11.63 | q.value = 5.57e-05 |
| 4 | HHIP AC109811.4 | IG15_AR3 | (Mip5FuR) | (Mip101) | FC = -11.53 | q.value = 3.75e-05 |
| 5 | KRR1 AC011611.26 | IG35_AR3 | (Mip5FuR) | (Mip101) | FC = -10.86 | q.value = 0.0118 |
| 6 | C12orf63 AC007564.10 | IG42_AR3 | (Mip5FuR) | (Mip101) | FC = 10.81 | q.value = 0.0119 |
| 7 | HHIP AC109811.4 | IG15_AR2 | (Mip5FuR) | (Mip101) | FC = -10.60 | q.value = 4.57e-05 |
| 8 | ANKRD11 SPG7 | IG22_AR7 | (Mip5FuR) | (Mip101) | FC = 9.88 | q.value = 0.000131 |
| 9 | HHIP AC109811.4 | IG15_AR1 | (Mip5FuR) | (Mip101) | FC = -9.58 | q.value = 0.000267 |
| 10 | HHIP AC109811.4 | IG15_AR8 | (Mip5FuR) | (Mip101) | FC = -9.53 | q.value = 0.0172 |
| 11 | APPL2 NUAK1 | IG12_AR30 | (Mip5FuR) | (Mip101) | FC = 8.32 | q.value = 0.0119 |
| 12 | HHIP AC109811.4 | IG15_AR11 | (Mip5FuR) | (Mip101) | FC = -7.25 | q.value = 0.00973 |
| 13 | SNORD56 AC004968.1 | IG27_AR8 | (Mip5FuR) | (Mip101) | FC = -7.22 | q.value = 0.0332 |
| 14 | HHIP AC109811.4 | IG15_AR4 | (Mip5FuR) | (Mip101) | FC = -7.14 | q.value = 0.0042 |
| 15 | AL163953.1 AL138479.3 | IG23_AR1 | (Mip5FuR) | (Mip101) | FC = -7.14 | q.value = 0.0323 |
| 16 | ZNF689 PRR14 | IG37_AR5 | (Mip5FuR) | (Mip101) | FC = 6.66 | q.value = 0.0119 |
| 17 | HHIP AC109811.4 | IG15_AR13 | (Mip5FuR) | (Mip101) | FC = -6.56 | q.value = 0.0137 |
| 18 | HHIP AC109811.4 | IG15_AR7 | (Mip5FuR) | (Mip101) | FC = -6.40 | q.value = 0.0137 |
| 19 | RP4-644F6.3 LRRC8B | IG35_AR2 | (Mip5FuR) | (Mip101) | FC = 5.99 | q.value = 0.0323 |
| 20 | TSPAN12 ING3 | IG23_AR1 | (Mip5FuR) | (Mip101) | FC = -5.75 | q.value = 5.77e-06 |
| 21 | HHIP AC109811.4 | IG15_AR5 | (Mip5FuR) | (Mip101) | FC = -5.71 | q.value = 0.00583 |
| 22 | SPATA9 RHOBTB3 | IG35_AR10 | (Mip5FuR) | (Mip101) | FC = 5.70 | q.value = 0.00972 |
| 23 | SPATA9 RHOBTB3 | IG35_AR11 | (Mip5FuR) | (Mip101) | FC = 5.30 | q.value = 0.0323 |
| 24 | FHAD1 EFHD2 | IG39_AR5 | (Mip5FuR) | (Mip101) | FC = 4.80 | q.value = 0.0039 |
| 25 | FMN1 AC019278.6 | IG25_AR10 | (Mip5FuR) | (Mip101) | FC = -4.78 | q.value = 0.00698 |
| 26 | AC096643.2 SLC30A10 | IG2_AR3 | (Mip5FuR) | (Mip101) | FC = -4.51 | q.value = 0.0408 |
| 27 | AC015804.14 TNRC6C | IG26_AR24 | (Mip5FuR) | (Mip101) | FC = -4.35 | q.value = 4.93e-08 |
| 28 | Y_RNA AIM1 | IG43_AR4 | (Mip5FuR) | (Mip101) | FC = -4.27 | q.value = 4.21e-11 |
| 29 | DLG1 BDH1 | IG26_AR1 | (Mip5FuR) | (Mip101) | FC = 3.78 | q.value = 0.00501 |
| 30 | 5S_rRNA CCNO | IG35_AR6 | (Mip5FuR) | (Mip101) | FC = -3.77 | q.value = 0.0173 |
| 31 | AP001042.1 AF064859.2 | IG48_AR20 | (Mip5FuR) | (Mip101) | FC = -3.74 | q.value = 4.81e-05 |
| 32 | AFG3L2 C18orf43 | IG20_AR1 | (Mip5FuR) | (Mip101) | FC = 3.68 | q.value = 0.0359 |
| 33 | AC015804.14 TNRC6C | IG26_AR20 | (Mip5FuR) | (Mip101) | FC = -3.59 | q.value = 2.14e-08 |
| 34 | HERPUD2 AC007551.1 | IG18_AR1 | (Mip5FuR) | (Mip101) | FC = 3.58 | q.value = 0.00382 |
| 35 | EPHB3 MAGEF1 | IG12_AR6 | (Mip5FuR) | (Mip101) | FC = 3.40 | q.value = 1.98e-48 |
| 36 | AIM1L AL451139.40 | IG48_AR1 | (Mip5FuR) | (Mip101) | FC = -3.37 | q.value = 8.43e-06 |
| 37 | AC092580.1 AC011747.5 | IG35_AR73 | (Mip5FuR) | (Mip101) | FC = -3.27 | q.value = 1.23e-10 |
| 38 | GPRIN2 PPYR1 | IG34_AR6 | (Mip5FuR) | (Mip101) | FC = 3.16 | q.value = 6.75e-05 |
| 39 | MAGEF1 VPS8 | IG13_AR1 | (Mip5FuR) | (Mip101) | FC = 3.13 | q.value = 5.98e-10 |
| 40 | TNRC6C AC021593.15 | IG27_AR1 | (Mip5FuR) | (Mip101) | FC = -3.01 | q.value = 0.000508 |
| 41 | SHC3 CKS2 | IG9_AR9 | (Mip5FuR) | (Mip101) | FC = 3.01 | q.value = 3.75e-05 |
| 42 | 7SK MGAT5 | IG12_AR4 | (Mip5FuR) | (Mip101) | FC = 2.95 | q.value = 0.00143 |
| 43 | GPRIN2 PPYR1 | IG34_AR7 | (Mip5FuR) | (Mip101) | FC = 2.84 | q.value = 0.0424 |
| 44 | AC046143.20 TMEM44 | IG38_AR2 | (Mip5FuR) | (Mip101) | FC = 2.81 | q.value = 0.00184 |
| 45 | AC069513.28 hsa-mir-570 | IG48_AR2 | (Mip5FuR) | (Mip101) | FC = 2.81 | q.value = 6.61e-07 |
| 46 | RAB26 SNORD60 | IG44_AR3 | (Mip5FuR) | (Mip101) | FC = 2.81 | q.value = 2.55e-24 |
| 47 | C14orf167 AL136419.3 | IG49_AR1 | (Mip5FuR) | (Mip101) | FC = -2.80 | q.value = 0.0042 |
| 48 | AP000751.4 | IG1_AR6 | (Mip5FuR) | (Mip101) | FC = -2.78 | q.value = 2.91e-09 |
| 49 | C11orf70 YAP1 | IG5_AR3 | (Mip5FuR) | (Mip101) | FC = -2.73 | q.value = 0.0473 |
| 50 | RPS3 KLHL35 | IG4_AR10 | (Mip5FuR) | (Mip101) | FC = 2.71 | q.value = 0.0091 |
| 51 | SERPINB1 SERPINB9 | IG29_AR1 | (Mip5FuR) | (Mip101) | FC = -2.68 | q.value = 0.000382 |
| 52 | NCBP2 PIGZ | IG23_AR1 | (Mip5FuR) | (Mip101) | FC = 2.65 | q.value = 1.71e-12 |
| 53 | ATG5 Y_RNA | IG40_AR3 | (Mip5FuR) | (Mip101) | FC = -2.62 | q.value = 0.0229 |
| 54 | NUDT16P NUDT16 | IG29_AR7 | (Mip5FuR) | (Mip101) | FC = -2.57 | q.value = 0.00906 |
| 55 | SSU_rRNA_5 | IG1_AR1 | (Mip5FuR) | (Mip101) | FC = 2.57 | q.value = 4.67e-142 |
| 56 | hsa-mir-196a-1 PRAC | IG46_AR18 | (Mip5FuR) | (Mip101) | FC = -2.56 | q.value = 2.83e-15 |
| 57 | AC011475.6 ILF3 | IG45_AR1 | (Mip5FuR) | (Mip101) | FC = 2.54 | q.value = 6.75e-05 |
| 58 | FAM55B CADM1 | IG17_AR9 | (Mip5FuR) | (Mip101) | FC = -2.54 | q.value = 0.00166 |
| 59 | SNORD60 TRAF7 | IG45_AR1 | (Mip5FuR) | (Mip101) | FC = 2.50 | q.value = 1.05e-36 |
| 60 | hsa-mir-196a-1 PRAC | IG46_AR19 | (Mip5FuR) | (Mip101) | FC = -2.50 | q.value = 2.1e-13 |
| 61 | CERK TBC1D22A | IG12_AR3 | (Mip5FuR) | (Mip101) | FC = 2.49 | q.value = 0.00143 |
| 62 | PF4 PPBP | IG46_AR1 | (Mip5FuR) | (Mip101) | FC = -2.49 | q.value = 0.0283 |
| 63 | AC096742.3 TACR3 | IG30_AR7 | (Mip5FuR) | (Mip101) | FC = -2.45 | q.value = 0.0373 |
| 64 | AP001042.1 AF064859.2 | IG48_AR22 | (Mip5FuR) | (Mip101) | FC = -2.39 | q.value = 0.0305 |
| 65 | PDCD6 AC010442.7 | IG6_AR6 | (Mip5FuR) | (Mip101) | FC = 2.39 | q.value = 0.0296 |
| 66 | AP000593.4 CLPB | IG13_AR4 | (Mip5FuR) | (Mip101) | FC = -2.36 | q.value = 0.0251 |
| 67 | APPL2 NUAK1 | IG12_AR5 | (Mip5FuR) | (Mip101) | FC = 2.36 | q.value = 0.0124 |
| 68 | GPRIN2 PPYR1 | IG34_AR4 | (Mip5FuR) | (Mip101) | FC = 2.33 | q.value = 0.0124 |
| 69 | hsa-mir-196a-1 PRAC | IG46_AR16 | (Mip5FuR) | (Mip101) | FC = -2.31 | q.value = 4.93e-08 |
| 70 | KIAA1913 Y_RNA | IG50_AR1 | (Mip5FuR) | (Mip101) | FC = -2.30 | q.value = 0.000185 |
| 71 | GREM1 FMN1 | IG24_AR12 | (Mip5FuR) | (Mip101) | FC = -2.29 | q.value = 0.0347 |
| 72 | AC069513.28 hsa-mir-570 | IG48_AR7 | (Mip5FuR) | (Mip101) | FC = 2.29 | q.value = 1.51e-06 |
| 73 | PCNX SNORD56 | IG26_AR15 | (Mip5FuR) | (Mip101) | FC = -2.29 | q.value = 0.0117 |
| 74 | KCNV1 AC104946.4 | IG20_AR2 | (Mip5FuR) | (Mip101) | FC = -2.28 | q.value = 4.59e-05 |
| 75 | hsa-mir-196a-1 PRAC | IG46_AR17 | (Mip5FuR) | (Mip101) | FC = -2.26 | q.value = 1.13e-08 |
| 76 | GPRIN2 PPYR1 | IG34_AR3 | (Mip5FuR) | (Mip101) | FC = 2.25 | q.value = 0.0172 |
| 77 | AP001042.1 AF064859.2 | IG48_AR21 | (Mip5FuR) | (Mip101) | FC = -2.24 | q.value = 0.0244 |
| 78 | GPRIN2 PPYR1 | IG34_AR5 | (Mip5FuR) | (Mip101) | FC = 2.22 | q.value = 0.0178 |
| 79 | S100A16 S100A14 | IG42_AR1 | (Mip5FuR) | (Mip101) | FC = -2.22 | q.value = 0.0339 |
| 80 | PTPRG C3orf14 | IG36_AR1 | (Mip5FuR) | (Mip101) | FC = -2.22 | q.value = 0.0221 |
| 81 | SNORND104 SNORA76 | IG14_AR1 | (Mip5FuR) | (Mip101) | FC = 2.19 | q.value = 5.55e-10 |
| 82 | AC005243.1 SLC39A11 | IG21_AR46 | (Mip5FuR) | (Mip101) | FC = 2.18 | q.value = 0.0347 |
| 83 | WRNIP1 SERPINB1 | IG28_AR4 | (Mip5FuR) | (Mip101) | FC = -2.16 | q.value = 1.19e-10 |
| 84 | hsa-mir-196a-1 PRAC | IG46_AR20 | (Mip5FuR) | (Mip101) | FC = -2.13 | q.value = 7.22e-07 |
| 85 | CTXN3 SLC12A2 | IG40_AR49 | (Mip5FuR) | (Mip101) | FC = 2.12 | q.value = 0.0427 |
| 86 | GPRIN2 PPYR1 | IG34_AR2 | (Mip5FuR) | (Mip101) | FC = 2.10 | q.value = 0.0427 |
| 87 | AC097015.2 TKT | IG35_AR2 | (Mip5FuR) | (Mip101) | FC = -2.10 | q.value = 0.000353 |
| 88 | PVRL3 CD96 | IG44_AR6 | (Mip5FuR) | (Mip101) | FC = 2.10 | q.value = 0.0364 |
| 89 | IFITM5 IFITM2 | IG9_AR3 | (Mip5FuR) | (Mip101) | FC = -2.09 | q.value = 1.14e-11 |
| 90 | U6 UBXN7 | IG12_AR14 | (Mip5FuR) | (Mip101) | FC = 2.09 | q.value = 0.00156 |
| 91 | PVRL3 CD96 | IG44_AR14 | (Mip5FuR) | (Mip101) | FC = 2.02 | q.value = 0.0137 |
| 92 | ABCB4 ABCB1 | IG7_AR4 | (Mip5FuR) | (Mip101) | FC = -2.02 | q.value = 0.0218 |
| 93 | HSPA1A HSPA1B | IG35_AR7 | (Mip5FuR) | (Mip101) | FC = -2.00 | q.value = 0.00336 |
| 94 | RANBP9 CCDC90A | IG35_AR10 | (Mip5FuR) | (Mip101) | FC = -2.00 | q.value = 0.0156 |