STRUCTURAL VARIATION
This section provides details of structural variations detected from de novo assembly of the tumour genome and transcriptome. Look in this section for details of the potentially relevant large-scale genomic rearrangements including gene fusions, duplications and deletions.
STRUCTURAL VARIATION
SUMMARY OF STRUCTURAL EVENTS
Structural Variants [Expressed Fusions]
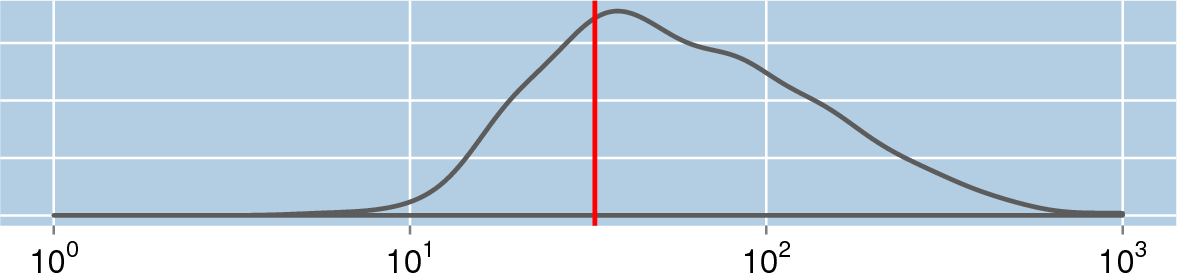
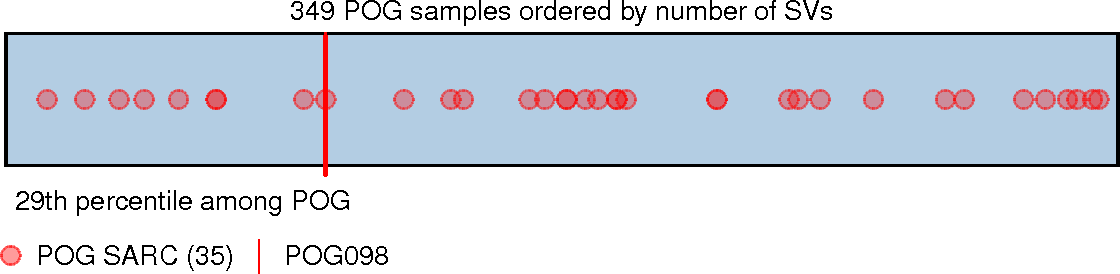
33[3]
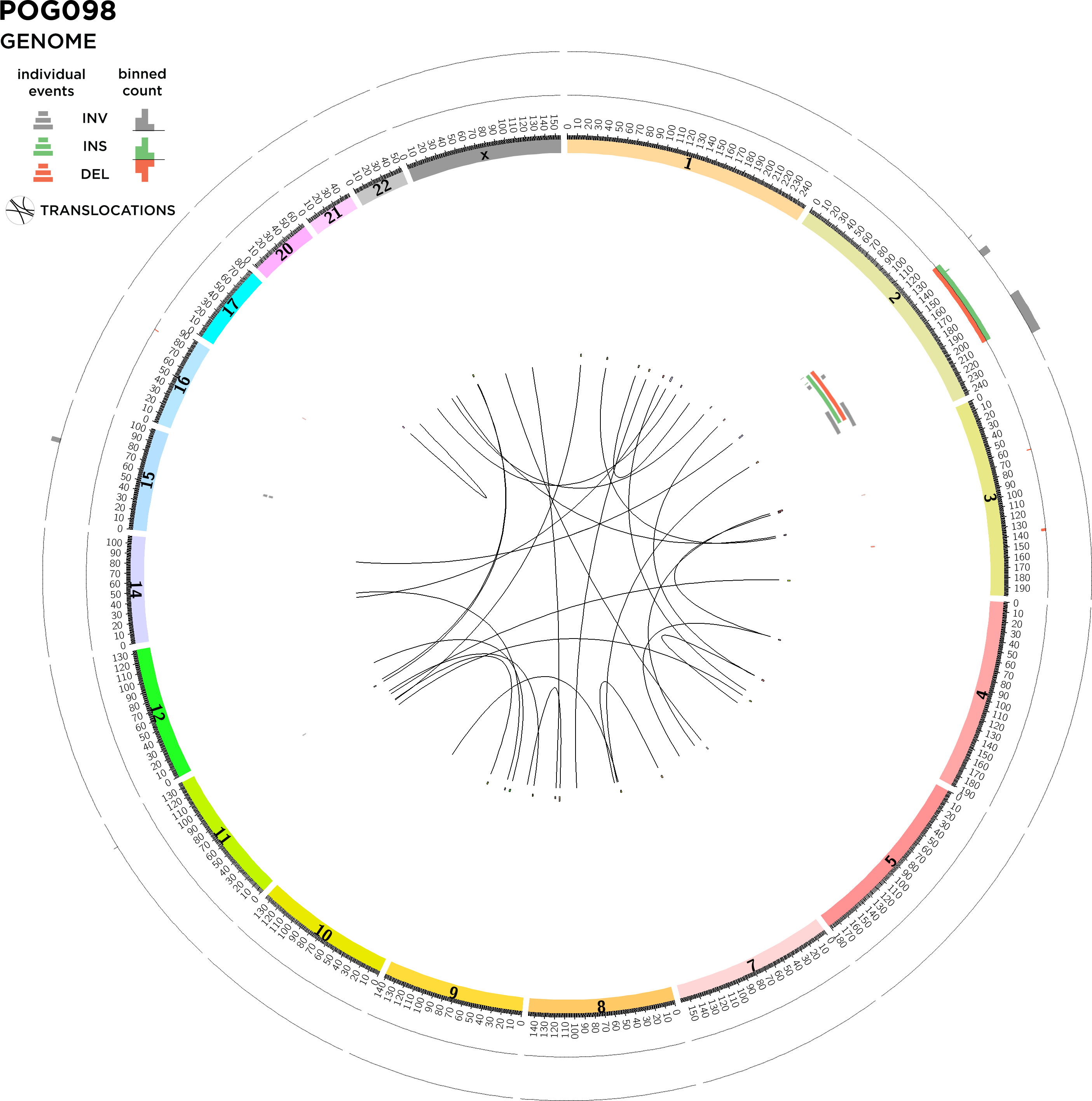
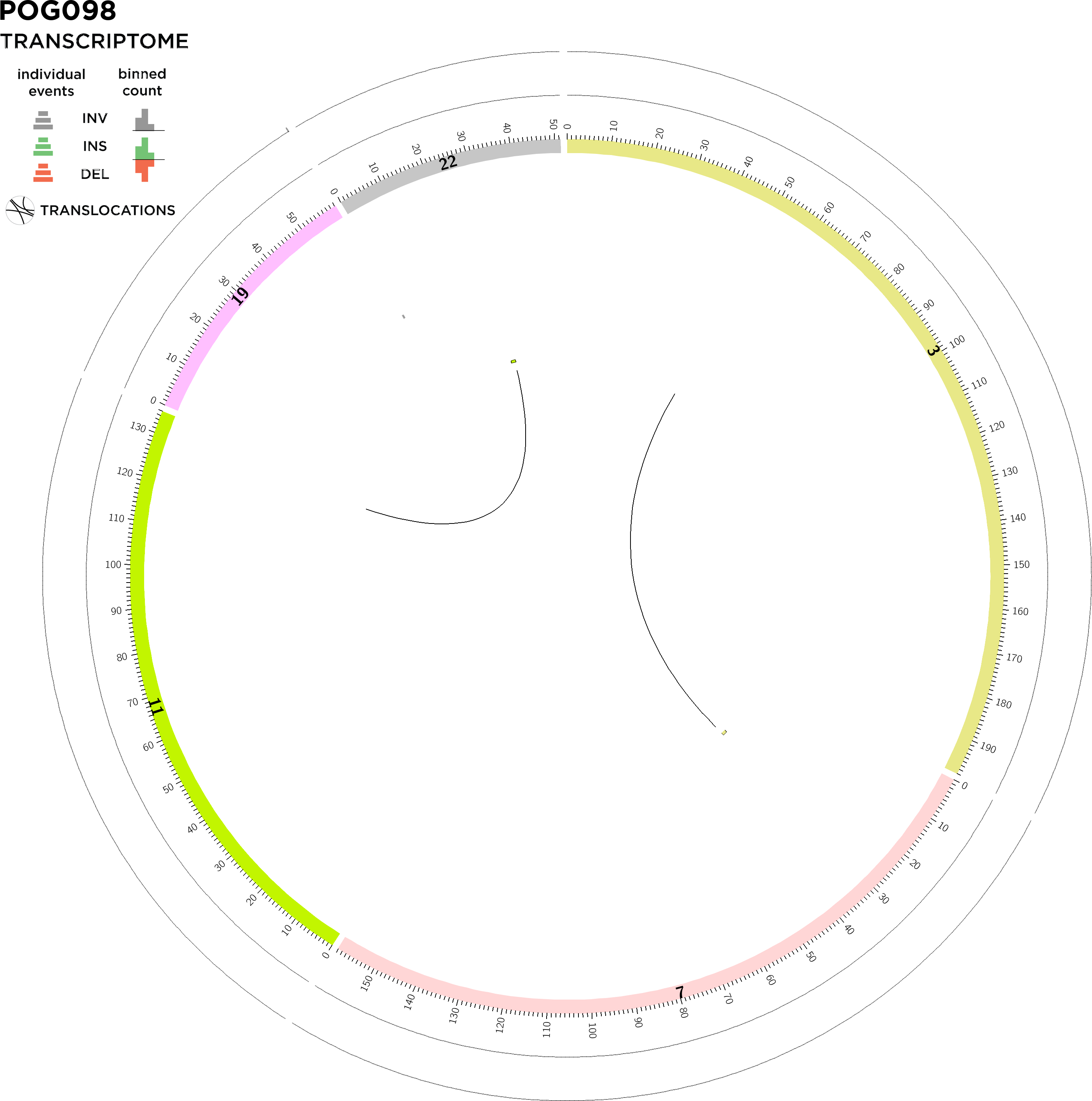
STRUCTURAL VARIATION: GENOMIC DETAILS
GENE FUSIONS OF POTENTIAL CLINICAL RELEVANCE WITH GENOME AND TRANSCRIPTOME SUPPORT
No gene fusions of potential clinical relevance were found from the analysis.
GENE FUSIONS OF PROGNOSTIC AND DIAGNOSTIC RELEVANCE
No gene fusions of prognostic and diagnostic relevance were found from the analysis.
GENE FUSIONS WITH BIOLOGICAL RELEVANCE
| Gene 5`::3` | Exons 5`/3` | Breakpoint | Event Type | Sample | Cytogenetic Description | RPKM 5`/3` | Fold Change vs. average | SARC %ile |
|---|---|---|---|---|---|---|---|---|
| EWSR1::FLI1 | e7:e4 | 22:29683123| 11:128675261 |
translocation | DNA/RNA | t(22;11)(q12.2;q24.3) | 10.96/6.37 | -1.4/-1.29 | 0/56 |
GENE FUSIONS WITH GENOME AND TRANSCRIPTOME SUPPORT
| Gene 5`::3` | Exons 5`/3` | Breakpoint | Event Type | Sample | Cytogenetic Description | RPKM 5`/3` | Fold Change vs. average | SARC %ile |
|---|---|---|---|---|---|---|---|---|
| TRIM28::NA | e2:e | 19:59056904| 19:59096255 |
inversion | DNA/RNA | i(19)(q13.43) | 93.05/na | 2.09/na | 56/na |
| WIPF3::IQCF4 | e:e | 7:29929023| 3:51851986 |
translocation | DNA/RNA | t(7;3)(p14.3;p21.2) | 15.39/na | 2.37/na | 100/na |
GENE FUSIONS FIGURES
Gene ID: EWSR1::FLI1
Type: translocation
Predicted: in-frame
| Position | HGNC Gene | Ensembl Gene | Ensembl Transcript | Exon | Strand | Breakpoint |
|---|---|---|---|---|---|---|
| 5` | EWSR1 | ENSG00000182944 | ENST00000397938 | e7 | 22:29683123 | |
| 3` | FLI1 | ENSG00000151702 | ENST00000525560 | e4 | 11:128675261 | |
| Putative Fusion-translocation event. The 5' (in the fusion) transcript EWSR1-001(ENST00000397938) from the gene EWSR1(ENSG00000182944) on the forward strand is
drawn top left with its corresponding breakpoint at 22:29683123. The 3' (in the fusion) transcript FLI1-004(ENST00000525560) from the gene FLI1(ENSG00000151702) on the forward strand is drawn top right with its corresponding breakpoint at 11:128675261. Exons are drawn to scale relative to other exons in the same drawing. Introns are scaled to make up approximately 1/4th of the final drawing. This is predicted to be an IN-frame fusion. Domain(s) featured in the above figure(s) are labelled by their various external identifiers as follows: Ets(PF00178); RRM_dom(PF00076); Znf_RanBP2(PF00641). |
||||||
Gene ID: TRIM28:NA
Type: inversion
Predicted: not determined
| Position | HGNC Gene | Ensembl Gene | Ensembl Transcript | Exon | Strand | Breakpoint |
|---|---|---|---|---|---|---|
| 5` | TRIM28 | ENSG00000130726 | ENST00000253024 | e2 | 19:59056904 | |
| 3` | NA | na | na | e | 19:59096255 | |
| Transcript TRIM28-201(ENST00000253024) from gene TRIM28(ENSG00000130726) on the forward strand. Breakpoint shown is 19:59056904. Exons are drawn to scale
relative to other exons in the same drawing. Introns are scaled to make up approximately 1/4th of the final drawing. Domain(s) featured in the above figure(s) are labelled by their various external identifiers as follows: Znf_B-box(PF00643); Znf_PHD-finger(PF00628). |
||||||

